235400 Caspase-3 Substrate I, Colorimetric - CAS 1177131-27-9 - Calbiochem
Colorimetric substrate for caspase-3 (Km = 9.7 µM) and related cysteine proteases.
More>> Colorimetric substrate for caspase-3 (Km = 9.7 µM) and related cysteine proteases. Less<<Synonyms: Ac-DEVD-pNA
Recommended Products
Overview
| Replacement Information |
|---|
| Product Information | |
|---|---|
| CAS number | 1177131-27-9 |
| ATP Competitive | N |
| Form | Lyophilized |
| Hill Formula | C₂₆H₃₄N₆O₁₃ |
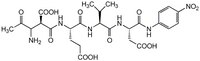
| Chemical formula | C₂₆H₃₄N₆O₁₃ |
| Reversible | N |
| Structure formula Image | |
| Quality Level | MQ100 |
| Applications |
|---|
| Biological Information | |
|---|---|
| Primary Target | Caspas-3 |
| Primary Target IC<sub>50</sub> | Km = 9.7 µM as colorimetric substrate for caspase-3 |
| Purity | ≥95% by HPLC |
| Physicochemical Information | |
|---|---|
| Cell permeable | N |
| Peptide Sequence | Ac-Asp-Glu-Val-Asp-pNA |
| Dimensions |
|---|
| Materials Information |
|---|
| Toxicological Information |
|---|
| Safety Information according to GHS |
|---|
| Safety Information |
|---|
| Product Usage Statements |
|---|
| Packaging Information |
|---|
| Transport Information |
|---|
| Supplemental Information |
|---|
| Specifications |
|---|
| Global Trade Item Number | |
|---|---|
| Catalogue Number | GTIN |
| 235400 | 0 |
Documentation
Caspase-3 Substrate I, Colorimetric - CAS 1177131-27-9 - Calbiochem MSDS
| Title |
|---|
Caspase-3 Substrate I, Colorimetric - CAS 1177131-27-9 - Calbiochem Certificates of Analysis
| Title | Lot Number |
|---|---|
| 235400 |
References
| Reference overview |
|---|
| Cardone, M.H., et al. 1998. Science 282, 1318. Thornberry, N.A., and Lazebnik, Y. 1998. Science 281, 1312. Nicholson, D.W., et al. 1995. Nature 376, 37. Lazebnik, Y.A., et al. 1994. Nature 371, 346. |