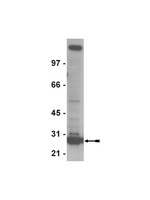
Impaired replication dynamics at the FRA3B common fragile site.
Aparna Palakodeti,Isabelle Lucas,Yanwen Jiang,David J Young,Anthony A Fernald,Theodore Karrison,Michelle M Le Beau
Human molecular genetics
19
2009
Zobrazit abstrakt
Chromosomal common fragile sites (CFSs) are genetically unstable regions of the genome that are induced by conditions that impair DNA replication. In this report, we show that treatment with the DNA polymerase inhibitor, aphidicolin (APH), slows the replication rate throughout S phase. To investigate the unusual sensitivity of CFSs to APH-induced replication stress, we examined replication dynamics within a 50 kb region of the most frequently expressed CFS, FRA3B. We mapped four origins of replication, ori 1-4, using two independent methods. In untreated cells, we detected significantly less newly replicated DNA at FRA3B ori 1-3, as compared with three control origins located within non-fragile regions (NCFSs). In APH-treated cells, all FRA3B and control origins tested were active; however, there was a significant increase of nascent strand DNA at the control origins and, to a lesser extent, at the FRA3B ori 1-3. On the basis of these observations and the theoretical modeling of the nascent strand abundance assay developed in this study, we hypothesize that CFS origins may be less efficient, and that APH treatment slows replication fork movement near these origins to a greater extent, resulting in impaired DNA replication and, ultimately, leading to the genetic instability characteristic of CFSs. Celý text článku | 19815620
 |
Complex protein-DNA dynamics at the latent origin of DNA replication of Epstein-Barr virus.
Ritzi, Marion, et al.
J. Cell. Sci., 116: 3971-84 (2003)
2003
Zobrazit abstrakt
The sequential binding of the origin recognition complex (ORC), Cdc6p and the minichromosome maintenance proteins (MCM2-7) mediates replication competence at eukaryotic origins of DNA replication. The latent origin of Epstein-Barr virus, oriP, is a viral origin known to recruit ORC. OriP also binds EBNA1, a virally encoded protein that lacks any activity predicted to be required for replication initiation. Here, we used chromatin immunoprecipitation and chromatin binding to compare the cell-cycle-dependent binding of pre-RC components and EBNA1 to oriP and to global cellular chromatin. Prereplicative-complex components such as the Mcm2p-Mcm7p proteins and HsOrc1p are regulated in a cell-cycle-dependent fashion, whereas other HsOrc subunits and EBNA1 remain constantly bound. In addition, HsOrc1p becomes sensitive to the 26S proteasome after release from DNA during S phase. These results show that the complex protein-DNA dynamics at the viral oriP are synchronized with the cell division cycle. Chromatin-binding and chromatin-immunoprecipitation experiments on G0 arrested cells indicated that the ORC core complex (ORC2-5) and EBNA1 remain bound to chromatin and oriP. HsOrc6p and the MCM2-7 complex are released in resting cells. HsOrc1p is partly liberated from chromatin. Our data suggest that origins remain marked in resting cells by the ORC core complex to ensure a rapid and regulated reentry into the cell cycle. These findings indicate that HsOrc is a dynamic complex and that its DNA binding activity is regulated differently in the various stages of the cell cycle. | 12953058
 |