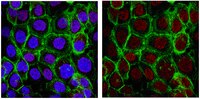
Foxa2 acts as a co-activator potentiating expression of the Nurr1-induced DA phenotype via epigenetic regulation.
Yi, SH; He, XB; Rhee, YH; Park, CH; Takizawa, T; Nakashima, K; Lee, SH
Development (Cambridge, England)
141
761-72
2014
Pokaż streszczenie
Understanding how dopamine (DA) phenotypes are acquired in midbrain DA (mDA) neuron development is important for bioassays and cell replacement therapy for mDA neuron-associated disorders. Here, we demonstrate a feed-forward mechanism of mDA neuron development involving Nurr1 and Foxa2. Nurr1 acts as a transcription factor for DA phenotype gene expression. However, Nurr1-mediated DA gene expression was inactivated by forming a protein complex with CoREST, and then recruiting histone deacetylase 1 (Hdac1), an enzyme catalyzing histone deacetylation, to DA gene promoters. Co-expression of Nurr1 and Foxa2 was established in mDA neuron precursor cells by a positive cross-regulatory loop. In the presence of Foxa2, the Nurr1-CoREST interaction was diminished (by competitive formation of the Nurr1-Foxa2 activator complex), and CoREST-Hdac1 proteins were less enriched in DA gene promoters. Consequently, histone 3 acetylation (H3Ac), which is responsible for open chromatin structures, was strikingly increased at DA phenotype gene promoters. These data establish the interplay of Nurr1 and Foxa2 as the crucial determinant for DA phenotype acquisition during mDA neuron development. | Immunofluorescence | 24496614
 |
DNA methyltransferase 3b preferentially associates with condensed chromatin.
Kashiwagi, K; Nimura, K; Ura, K; Kaneda, Y
Nucleic acids research
39
874-88
2010
Pokaż streszczenie
In mammals, DNA methylation is catalyzed by DNA methyltransferases (DNMTs) encoded by Dnmt1, Dnmt3a and Dnmt3b. Since, the mechanisms of regulation of Dnmts are still largely unknown, the physical interaction between Dnmt3b and chromatin was investigated in vivo and in vitro. In embryonic stem cell nuclei, Dnmt3b preferentially associated with histone H1-containing heterochromatin without any significant enrichment of silent-specific histone methylation. Recombinant Dnmt3b preferentially associated with nucleosomal DNA rather than naked DNA. Incorporation of histone H1 into nucleosomal arrays promoted the association of Dnmt3b with chromatin, whereas histone acetylation reduced Dnmt3b binding in vitro. In addition, Dnmt3b associated with histone deacetylase SirT1 in the nuclease resistant chromatin. These findings suggest that Dnmt3b is preferentially recruited into hypoacetylated and condensed chromatin. We propose that Dnmt3b is a 'reader' of higher-order chromatin structure leading to gene silencing through DNA methylation. | Western Blotting | 20923784
 |
Fgfr2 is required for the development of the medial prefrontal cortex and its connections with limbic circuits.
Stevens, HE; Smith, KM; Maragnoli, ME; Fagel, D; Borok, E; Shanabrough, M; Horvath, TL; Vaccarino, FM
The Journal of neuroscience : the official journal of the Society for Neuroscience
30
5590-602
2009
Pokaż streszczenie
To understand the role of specific fibroblast growth factor receptors (FGFRs) in cortical development, we conditionally inactivated Fgfr2 or both Fgfr1 and Fgfr2 [Fgfr2 conditional knock-out (cKO) or double knock-out mice, respectively] in radial glial cells of the dorsal telencephalon. Fgfr1 and Fgfr2 are necessary for the attainment of a normal number of excitatory neurons in the cerebral cortex. The action of FGF receptors appears to be through increasing self-renewal of neuronal precursors within the ventricular zone. Volume measurements, assessments of excitatory neuron number, and areal marker expression suggested that the proper formation of the medial prefrontal cortex (mPFC) depends on the function of Fgfr2, whereas Fgfr1 together with Fgfr2 control excitatory cortical neuron development within the entire cerebral cortex. Fgfr2 cKO mice had fewer and smaller glutamate synaptic terminals in the bed nuclei of the stria terminalis (BST), a projection area for mPFC cortical neurons. Furthermore, Fgfr2 cKO mice showed secondary decreases in GABAergic neurons in the BST and septum. These data demonstrate that FGFR2 signaling expands the number of excitatory neurons in the mPFC and secondarily influences target neurons in subcortical stations of the limbic system. Pełny tekst artykułu | | 20410112
 |
Lsh, a modulator of CpG methylation, is crucial for normal histone methylation.
Yan, Q; Huang, J; Fan, T; Zhu, H; Muegge, K
The EMBO journal
22
5154-62
2003
Pokaż streszczenie
Methylation of histone tails and CpG methylation are involved in determining heterochromatin structure, but their cause and effect relationship has not been resolved as yet in mammals. Here we report that Lsh, a member of the SNF2 chromatin remodeling family, controls both types of epigenetic modifications. Lsh has been shown to be associated with pericentromeric heterochromatin and to be required for normal CpG methylation at pericentromeric sequences. Loss of Lsh, in Lsh-deficient mice, results in accumulation of di- and tri-methylated histone 3 at lysine 4 (H3-K4me) at pericentromeric DNA and other repetitive sequences. In contrast, di- or tri-methylation of H3-K9 and distribution of HP1 appear unchanged after Lsh deletion, suggesting independent regulatory mechanisms for H3-K4 or K9 methylation. Experimental DNA demethylation with 5'-azacytidine results in a similar increase of H3-K4me. These results support the model that loss of CpG methylation caused by Lsh deficiency antecedes elevation of H3-K4me. Thus, Lsh is crucial for the formation of normal heterochromatin, implying a functional role for Lsh in the regulation of transcription and mitosis. | Western Blotting | 14517253
 |
Defective flap endonuclease 1 activity in mammalian cells is associated with impaired DNA repair and prolonged S phase delay
Shibata, Y. and Nakamura, T.
J Biol Chem, 277:746-54 (2002)
2002
| Immunoblotting (Western) | 11687589
 |
Adenosine reverses a preestablished CCl4-induced micronodular cirrhosis through enhancing collagenolytic activity and stimulating hepatocyte cell proliferation in rats
Hernandez-Munoz, R., et al
Hepatology, 34:677-87 (2001)
2001
| Immunoblotting (Western) | 11584363
 |
Proliferating cell nuclear antigen is required for DNA excision repair.
Shivji, K K, et al.
Cell, 69: 367-74 (1992)
1992
Pokaż streszczenie
Fractionation of extracts from human cell lines allows nucleotide excision repair of damaged DNA to be resolved into discrete incision and polymerization stages. Generation of incised intermediates depends on the XP-A protein, a polypeptide that recognizes sites of damaged DNA, and on the human single-stranded DNA-binding protein HSSB. The proliferating cell nuclear antigen (PCNA) is required for the DNA synthesis that converts the nicked intermediates to completed repair events. This need for PCNA implies that repair synthesis is carried out by DNA polymerase delta or epsilon. The ability to visualize repair intermediates in the absence of PCNA facilitates dissection of the multiprotein reaction that leads to incision of damaged DNA in a major pathway of cellular defense against mutagens. | | 1348971
 |
Monoclonal antibody analysis of the proliferating cell nuclear antigen (PCNA). Structural conservation and the detection of a nucleolar form.
Waseem, N H and Lane, D P
J. Cell. Sci., 96 ( Pt 1): 121-9 (1990)
1990
Pokaż streszczenie
The proliferating cell nuclear antigen, PCNA, has recently been identified as the polymerase delta accessory protein. PCNA is essential for cellular DNA synthesis and is also required for the in vitro replication of simian virus 40 (SV40) DNA where it acts to coordinate leading and lagging strand synthesis at the replication fork. The cDNA for rat PCNA was cloned into a series of bacterial expression vectors and the resulting protein used to immunize mice. Eleven new monoclonal antibodies to PCNA have been isolated and characterized. Some of the antibodies recognize epitopes conserved from man to fission yeast. Immunocytochemical analysis of primate epithelial cell lines showed that the antibodies recognized antigenically distinct forms of PCNA and that these forms were localized to different compartments of the nucleus. One antibody reacted exclusively with PCNA in the nucleolus. These results suggest that the PCNA protein may fulfil several separate roles in the cell nucleus associated with changes in its antigenic structure. | | 1695635
 |
Mechanism of elongation of primed DNA by DNA polymerase delta, proliferating cell nuclear antigen, and activator 1.
Lee, S H and Hurwitz, J
Proc. Natl. Acad. Sci. U.S.A., 87: 5672-6 (1990)
1990
Pokaż streszczenie
In the presence of a single-stranded-DNA-binding protein (SSB), the elongation of primed DNA templates by DNA polymerase delta (pol delta) is dependent on ATP and two protein factors, activator 1 (A1) and proliferating cell nuclear antigen (PCNA). We have examined the interaction of these proteins with (dA)4500.(dT)12-18 by measuring their ability to form stable complexes with this DNA. In the presence of ATP, A1, PCNA, and pol delta formed a stable complex with DNA that could be isolated by gel filtration. Incubation of the isolated complex with dTTP resulted in the synthesis of poly(dT). While ATP was required for the formation of this complex, it was not required for the subsequent elongation of DNA. The temporal requirements for complex formation were determined. A1 was found to bind first, followed by the ATP-dependent addition of PCNA to the A1.DNA complex, while pol delta was added last. Each of these complexes could be isolated by gel filtration, indicating that they possessed a high degree of stability. The binding of PCNA to the A1-SSB-coated primed DNA occurred with adenosine 5'-[gamma-thio]triphosphate as well as ATP. However, the binding of pol delta to the PCNA.A1-DNA complex was observed only when the latter complex was formed in the presence of ATP. The complete complex was formed after incubation at 37 degrees C for 2 min, whereas no complex was detected after incubation at 0 degree C. These results indicate that these proteins act in a manner analogous to the accessory proteins that play critical roles in the elongation reaction catalyzed by T4 phage DNA polymerase and Escherichia coli DNA polymerase III. | | 1974050
 |