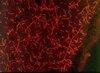
Co-visualization of DNA damage and ion traversals in live mammalian cells using a fluorescent nuclear track detector.
Kodaira, S; Konishi, T; Kobayashi, A; Maeda, T; Ahmad, TA; Yang, G; Akselrod, MS; Furusawa, Y; Uchihori, Y
Journal of radiation research
56
360-5
2015
요약 표시
The geometric locations of ion traversals in mammalian cells constitute important information in the study of heavy ion-induced biological effect. Single ion traversal through a cellular nucleus produces complex and massive DNA damage at a nanometer level, leading to cell inactivation, mutations and transformation. We present a novel approach that uses a fluorescent nuclear track detector (FNTD) for the simultaneous detection of the geometrical images of ion traversals and DNA damage in single cells using confocal microscopy. HT1080 or HT1080-53BP1-GFP cells were cultured on the surface of a FNTD and exposed to 5.1-MeV/n neon ions. The positions of the ion traversals were obtained as fluorescent images of a FNTD. Localized DNA damage in cells was identified as fluorescent spots of γ-H2AX or 53BP1-GFP. These track images and images of damaged DNA were obtained in a short time using a confocal laser scanning microscope. The geometrical distribution of DNA damage indicated by fluorescent γ-H2AX spots in fixed cells or fluorescent 53BP1-GFP spots in living cells was found to correlate well with the distribution of the ion traversals. This method will be useful for evaluating the number of ion hits on individual cells, not only for micro-beam but also for random-beam experiments. | | | 25324538
 |
A functional screen identifies miRNAs that inhibit DNA repair and sensitize prostate cancer cells to ionizing radiation.
Hatano, K; Kumar, B; Zhang, Y; Coulter, JB; Hedayati, M; Mears, B; Ni, X; Kudrolli, TA; Chowdhury, WH; Rodriguez, R; DeWeese, TL; Lupold, SE
Nucleic acids research
43
4075-86
2015
요약 표시
MicroRNAs (miRNAs) have been implicated in DNA repair pathways through transcriptional responses to DNA damaging agents or through predicted miRNA regulation of DNA repair genes. We hypothesized that additional DNA damage regulating miRNAs could be identified by screening a library of 810 miRNA mimetics for the ability to alter cellular sensitivity to ionizing radiation (IR). A prostate cancer Metridia luciferase cell model was applied to examine the effects of individual miRNAs on IR sensitivity. A large percentage of miRNA mimetics were found to increase cellular sensitivity to IR, while a smaller percentage were protective. Two of the most potent IR sensitizing miRNAs, miR-890 and miR-744-3p, significantly delayed IR induced DNA damage repair. Both miRNAs inhibited the expression of multiple components of DNA damage response and DNA repair. miR-890 directly targeted MAD2L2, as well as WEE1 and XPC, where miR-744-3p directly targeted RAD23B. Knock-down of individual miR-890 targets by siRNA was not sufficient to ablate miR-890 radiosensitization, signifying that miR-890 functions by regulating multiple DNA repair genes. Intratumoral delivery of miR-890 mimetics prior to IR therapy significantly enhanced IR therapeutic efficacy. These results reveal novel miRNA regulation of DNA repair and identify miR-890 as a potent IR sensitizing agent. | | | 25845598
 |
MK3 modulation affects BMI1-dependent and independent cell cycle check-points.
Prickaerts, P; Niessen, HE; Dahlmans, VE; Spaapen, F; Salvaing, J; Vanhove, J; Geijselaers, C; Bartels, SJ; Partouns, I; Neumann, D; Speel, EJ; Takihara, Y; Wouters, BG; Voncken, JW
PloS one
10
e0118840
2015
요약 표시
Although the MK3 gene was originally found deleted in some cancers, it is highly expressed in others. The relevance of MK3 for oncogenesis is currently not clear. We recently reported that MK3 controls ERK activity via a negative feedback mechanism. This prompted us to investigate a potential role for MK3 in cell proliferation. We here show that overexpression of MK3 induces a proliferative arrest in normal diploid human fibroblasts, characterized by enhanced expression of replication stress- and senescence-associated markers. Surprisingly, MK3 depletion evokes similar senescence characteristics in the fibroblast model. We previously identified MK3 as a binding partner of Polycomb Repressive Complex 1 (PRC1) proteins. In the current study we show that MK3 overexpression results in reduced cellular EZH2 levels and concomitant loss of epigenetic H3K27me3-marking and PRC1/chromatin-occupation at the CDKN2A/INK4A locus. In agreement with this, the PRC1 oncoprotein BMI1, but not the PCR2 protein EZH2, bypasses MK3-induced senescence in fibroblasts and suppresses P16INK4A expression. In contrast, BMI1 does not rescue the MK3 loss-of-function phenotype, suggesting the involvement of multiple different checkpoints in gain and loss of MK3 function. Notably, MK3 ablation enhances proliferation in two different cancer cells. Finally, the fibroblast model was used to evaluate the effect of potential tumorigenic MK3 driver-mutations on cell proliferation and M/SAPK signaling imbalance. Taken together, our findings support a role for MK3 in control of proliferation and replicative life-span, in part through concerted action with BMI1, and suggest that the effect of MK3 modulation or mutation on M/SAPK signaling and, ultimately, proliferation, is cell context-dependent. | | | 25853770
 |
Loss of INPP4B causes a DNA repair defect through loss of BRCA1, ATM and ATR and can be targeted with PARP inhibitor treatment.
Ip, LR; Poulogiannis, G; Viciano, FC; Sasaki, J; Kofuji, S; Spanswick, VJ; Hochhauser, D; Hartley, JA; Sasaki, T; Gewinner, CA
Oncotarget
6
10548-62
2015
요약 표시
Treatment options for ovarian cancer patients remain limited and overall survival is less than 50% despite recent clinical advances. The lipid phosphatase inositol polyphosphate 4-phosphatase type II (INPP4B) has been described as a tumor suppressor in the PI3K/Akt pathway with loss of expression found most pronounced in breast, ovarian cancer and melanoma. Using microarray technology we identified a DNA repair defect in INPP4B-deficient cells, which we further characterized by comet assays and quantification of γH2AX, RAD51 and 53BP1 foci formation. INPP4B loss resulted in significantly increased sensitivity towards PARP inhibition, comparable to loss of BRCA1 in two- and three-dimensional in vitro models, as well as in in vivo xenograft models. Mechanistically, we discovered that INPP4B forms a protein complex with the key players of DNA repair, ATR and BRCA1, in GST pulldown and 293T overexpression assays, and INPP4B loss affects BRCA1, ATM and ATR protein stability resulting in the observed DNA repair defect. Given that INPP4B loss has been found in 40% of ovarian cancer patients, this study provides the rationale for establishing INPP4B as a biomarker of PARP inhibitor response, and consequently offers novel therapeutic options for a significant subset of patients. Loss of the tumor suppressor inositol polyphosphate 4-phosphatase type II (INPP4B) results in a DNA repair defect due to concomitant loss of BRCA1, ATR and ATM and can be therapeutically targeted with PARP inhibitors. | Western Blotting | | 25868852
 |
XPC inhibits NSCLC cell proliferation and migration by enhancing E-Cadherin expression.
Cui, T; Srivastava, AK; Han, C; Yang, L; Zhao, R; Zou, N; Qu, M; Duan, W; Zhang, X; Wang, QE
Oncotarget
6
10060-72
2015
요약 표시
Xeroderma pigmentosum complementation group C (XPC) protein is an important DNA damage recognition factor in nucleotide excision repair. Deletion of XPC is associated with early stages of human lung carcinogenesis, and reduced XPC mRNA levels predict poor patient outcome for non-small cell lung cancer (NSCLC). However, the mechanisms linking loss of XPC expression and poor prognosis in lung cancer are still unclear. Here, we report evidence that XPC silencing drives proliferation and migration of NSCLC cells by down-regulating E-Cadherin. XPC knockdown enhanced proliferation and migration while decreasing E-Cadherin expression in NSCLC cells with an epithelial phenotype. Restoration of E-Cadherin in these cells suppressed XPC knockdown-induced cell growth both in vitro and in vivo. Mechanistic studies showed that the loss of XPC repressed E-Cadherin expression by activating the ERK pathway and upregulating Snail expression. Our findings indicate that XPC silencing-induced reduction of E-Cadherin expression contributes, at least in part, to the poor outcome of NSCLC patients with low XPC expression. | | | 25871391
 |
Human papillomaviruses activate and recruit SMC1 cohesin proteins for the differentiation-dependent life cycle through association with CTCF insulators.
Mehta, K; Gunasekharan, V; Satsuka, A; Laimins, LA
PLoS pathogens
11
e1004763
2015
요약 표시
Human papillomaviruses infect stratified epithelia and link their productive life cycle to the differentiation state of the host cell. Productive viral replication or amplification is restricted to highly differentiated suprabasal cells and is dependent on the activation of the ATM DNA damage pathway. The ATM pathway has three arms that can act independently of one another. One arm is centered on p53, another on CHK2 and a third on SMC1/NBS1 proteins. A role for CHK2 in HPV genome amplification has been demonstrated but it was unclear what other factors provided important activities. The cohesin protein, SMC1, is necessary for sister chromatid association prior to mitosis. In addition the phosphorylated form of SMC1 plays a critical role together with NBS1 in the ATM DNA damage response. In normal cells, SMC1 becomes phosphorylated in response to radiation, however, in HPV positive cells our studies demonstrate that it is constitutively activated. Furthermore, pSMC1 is found localized in distinct nuclear foci in complexes with γ-H2AX, and CHK2 and bound to HPV DNA. Importantly, knockdown of SMC1 blocks differentiation-dependent genome amplification. pSMC1 forms complexes with the insulator transcription factor CTCF and our studies show that these factors bind to conserved sequence motifs in the L2 late region of HPV 31. Similar motifs are found in most HPV types. Knockdown of CTCF with shRNAs blocks genome amplification and mutation of the CTCF binding motifs in the L2 open reading frame inhibits stable maintenance of viral episomes in undifferentiated cells as well as amplification of genomes upon differentiation. These findings suggest a model in which SMC1 factors are constitutively activated in HPV positive cells and recruited to viral genomes through complex formation with CTCF to facilitate genome amplification. Our findings identify both SMC1 and CTCF as critical regulators of the differentiation-dependent life cycle of high-risk human papillomaviruses. | | | 25875106
 |
Specific uptake and genotoxicity induced by polystyrene nanobeads with distinct surface chemistry on human lung epithelial cells and macrophages.
Paget, V; Dekali, S; Kortulewski, T; Grall, R; Gamez, C; Blazy, K; Aguerre-Chariol, O; Chevillard, S; Braun, A; Rat, P; Lacroix, G
PloS one
10
e0123297
2015
요약 표시
Nanoparticle surface chemistry is known to play a crucial role in interactions with cells and their related cytotoxic effects. As inhalation is a major route of exposure to nanoparticles, we studied specific uptake and damages of well-characterized fluorescent 50 nm polystyrene (PS) nanobeads harboring different functionalized surfaces (non-functionalized, carboxylated and aminated) on pulmonary epithelial cells and macrophages (Calu-3 and THP-1 cell lines respectively). Cytotoxicity of in mass dye-labeled functionalized PS nanobeads was assessed by xCELLigence system and alamarBlue viability assay. Nanobeads-cells interactions were studied by video-microscopy, flow cytometry and also confocal microscopy. Finally ROS generation was assessed by glutathione depletion dosages and genotoxicity was assessed by γ-H2Ax foci detection, which is considered as the most sensitive technique for studying DNA double strand breaks. The uptake kinetic was different for each cell line. All nanobeads were partly adsorbed and internalized, then released by Calu-3 cells, while THP-1 macrophages quickly incorporated all nanobeads which were located in the cytoplasm rather than in the nuclei. In parallel, the genotoxicity study reported that only aminated nanobeads significantly increased DNA damages in association with a strong depletion of reduced glutathione in both cell lines. We showed that for similar nanoparticle concentrations and sizes, aminated polystyrene nanobeads were more cytotoxic and genotoxic than unmodified and carboxylated ones on both cell lines. Interestingly, aminated polystyrene nanobeads induced similar cytotoxic and genotoxic effects on Calu-3 epithelial cells and THP-1 macrophages, for all levels of intracellular nanoparticles tested. Our results strongly support the primordial role of nanoparticles surface chemistry on cellular uptake and related biological effects. Moreover our data clearly show that nanoparticle internalization and observed adverse effects are not necessarily associated. | | | 25875304
 |
A localized nucleolar DNA damage response facilitates recruitment of the homology-directed repair machinery independent of cell cycle stage.
van Sluis, M; McStay, B
Genes & development
29
1151-63
2015
요약 표시
DNA double-strand breaks (DSBs) are repaired by two main pathways: nonhomologous end-joining and homologous recombination (HR). Repair pathway choice is thought to be determined by cell cycle timing and chromatin context. Nucleoli, prominent nuclear subdomains and sites of ribosome biogenesis, form around nucleolar organizer regions (NORs) that contain rDNA arrays located on human acrocentric chromosome p-arms. Actively transcribed rDNA repeats are positioned within the interior of the nucleolus, whereas sequences proximal and distal to NORs are packaged as heterochromatin located at the nucleolar periphery. NORs provide an opportunity to investigate the DSB response at highly transcribed, repetitive, and essential loci. Targeted introduction of DSBs into rDNA, but not abutting sequences, results in ATM-dependent inhibition of their transcription by RNA polymerase I. This is coupled with movement of rDNA from the nucleolar interior to anchoring points at the periphery. Reorganization renders rDNA accessible to repair factors normally excluded from nucleoli. Importantly, DSBs within rDNA recruit the HR machinery throughout the cell cycle. Additionally, unscheduled DNA synthesis, consistent with HR at damaged NORs, can be observed in G1 cells. These results suggest that HR can be templated in cis and suggest a role for chromosomal context in the maintenance of NOR genomic stability. | | | 26019174
 |
HMGB1 facilitates repair of mitochondrial DNA damage and extends the lifespan of mutant ataxin-1 knock-in mice.
Ito, H; Fujita, K; Tagawa, K; Chen, X; Homma, H; Sasabe, T; Shimizu, J; Shimizu, S; Tamura, T; Muramatsu, S; Okazawa, H
EMBO molecular medicine
7
78-101
2015
요약 표시
Mutant ataxin-1 (Atxn1), which causes spinocerebellar ataxia type 1 (SCA1), binds to and impairs the function of high-mobility group box 1 (HMGB1), a crucial nuclear protein that regulates DNA architectural changes essential for DNA damage repair and transcription. In this study, we established that transgenic or virus vector-mediated complementation with HMGB1 ameliorates motor dysfunction and prolongs lifespan in mutant Atxn1 knock-in (Atxn1-KI) mice. We identified mitochondrial DNA damage repair by HMGB1 as a novel molecular basis for this effect, in addition to the mechanisms already associated with HMGB1 function, such as nuclear DNA damage repair and nuclear transcription. The dysfunction and the improvement of mitochondrial DNA damage repair functions are tightly associated with the exacerbation and rescue, respectively, of symptoms, supporting the involvement of mitochondrial DNA quality control by HMGB1 in SCA1 pathology. Moreover, we show that the rescue of Purkinje cell dendrites and dendritic spines by HMGB1 could be downstream effects. Although extracellular HMGB1 triggers inflammation mediated by Toll-like receptor and receptor for advanced glycation end products, upregulation of intracellular HMGB1 does not induce such side effects. Thus, viral delivery of HMGB1 is a candidate approach by which to modify the disease progression of SCA1 even after the onset. | | | 25510912
 |
NF-κB signaling mediates acquired resistance after PARP inhibition.
Nakagawa, Y; Sedukhina, AS; Okamoto, N; Nagasawa, S; Suzuki, N; Ohta, T; Hattori, H; Roche-Molina, M; Narváez, AJ; Jeyasekharan, AD; Bernal, JA; Sato, K
Oncotarget
6
3825-39
2015
요약 표시
PARP inhibitors are a class of promising anti-cancer drugs, with proven activity in BRCA mutant cancers. However, as with other targeted agents, treatment with PARP inhibitors generates acquired resistance within these tumors. The mechanism of this acquired resistance is poorly understood. We established cell lines that are resistant to PARP inhibitor by continuous treatment with the drug, and then used RNA sequencing to compare gene expression. Pathway analysis on the RNA sequencing data indicates that NF-κB signaling is preferentially up-regulated in PARP inhibitor-resistant cells, and that knockdown of core components in NF-κB signaling reverses the sensitivity to PARP inhibitor in resistant cells. Of therapeutic relevance, we show that PARP inhibitor-resistant cells are sensitive to an NF-κB inhibitor in comparison to their parental controls. Malignancies with up-regulation of NF-κB are sensitive to bortezomib, a proteasome inhibitor that is currently used in the clinic. We also show that treatment with bortezomib results in cell death in the PARP inhibitor-resistant cells, but not in parental cells. Therefore we propose that up-regulation of NF-κB signaling is a key mechanism underlying acquired resistance to PARP inhibition, and that NF-κB inhibition, or bortezomib are potentially effective anti-cancer agents after the acquisition of resistance to PARP inhibitors. | | | 25686825
 |