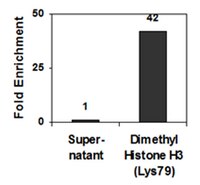
Methylation of histone H3 on lysine 79 associates with a group of replication origins and helps limit DNA replication once per cell cycle.
Fu, H; Maunakea, AK; Martin, MM; Huang, L; Zhang, Y; Ryan, M; Kim, R; Lin, CM; Zhao, K; Aladjem, MI
PLoS genetics
9
e1003542
2013
요약 표시
Mammalian DNA replication starts at distinct chromosomal sites in a tissue-specific pattern coordinated with transcription, but previous studies have not yet identified a chromatin modification that correlates with the initiation of DNA replication at particular genomic locations. Here we report that a distinct fraction of replication initiation sites in the human genome are associated with a high frequency of dimethylation of histone H3 lysine K79 (H3K79Me2). H3K79Me2-containing chromatin exhibited the highest genome-wide enrichment for replication initiation events observed for any chromatin modification examined thus far (23.39% of H3K79Me2 peaks were detected in regions adjacent to replication initiation events). The association of H3K79Me2 with replication initiation sites was independent and not synergistic with other chromatin modifications. H3K79 dimethylation exhibited wider distribution on chromatin during S-phase, but only regions with H3K79 methylation in G1 and G2 were enriched in replication initiation events. H3K79 was dimethylated in a region containing a functional replicator (a DNA sequence capable of initiating DNA replication), but the methylation was not evident in a mutant replicator that could not initiate replication. Depletion of DOT1L, the sole enzyme responsible for H3K79 methylation, triggered limited genomic over-replication although most cells could continue to proliferate and replicate DNA in the absence of methylated H3K79. Thus, prevention of H3K79 methylation might affect regulatory processes that modulate the order and timing of DNA replication. These data are consistent with the hypothesis that dimethylated H3K79 associates with some replication origins and marks replicated chromatin during S-phase to prevent re-replication and preserve genomic stability. | 23754963
 |
Coordinate changes in histone modifications, mRNA levels, and metabolite profiles in clonal INS-1 832/13 β-cells accompany functional adaptations to lipotoxicity.
Malmgren, S; Spégel, P; Danielsson, AP; Nagorny, CL; Andersson, LE; Nitert, MD; Ridderstråle, M; Mulder, H; Ling, C
The Journal of biological chemistry
288
11973-87
2013
요약 표시
Lipotoxicity is a presumed pathogenetic process whereby elevated circulating and stored lipids in type 2 diabetes cause pancreatic β-cell failure. To resolve the underlying molecular mechanisms, we exposed clonal INS-1 832/13 β-cells to palmitate for 48 h. We observed elevated basal insulin secretion but impaired glucose-stimulated insulin secretion in palmitate-exposed cells. Glucose utilization was unchanged, palmitate oxidation was increased, and oxygen consumption was impaired. Halting exposure of the clonal INS-1 832/13 β-cells to palmitate largely recovered all of the lipid-induced functional changes. Metabolite profiling revealed profound but reversible increases in cellular lipids. Glucose-induced increases in tricarboxylic acid cycle intermediates were attenuated by exposure to palmitate. Analysis of gene expression by microarray showed increased expression of 982 genes and decreased expression of 1032 genes after exposure to palmitate. Increases were seen in pathways for steroid biosynthesis, cell cycle, fatty acid metabolism, DNA replication, and biosynthesis of unsaturated fatty acids; decreases occurred in the aminoacyl-tRNA synthesis pathway. The activity of histone-modifying enzymes and histone modifications of differentially expressed genes were reversibly altered upon exposure to palmitate. Thus, Insig1, Lss, Peci, Idi1, Hmgcs1, and Casr were subject to epigenetic regulation. Our analyses demonstrate that coordinate changes in histone modifications, mRNA levels, and metabolite profiles accompanied functional adaptations of clonal β-cells to lipotoxicity. It is highly likely that these changes are pathogenetic, accounting for loss of glucose responsiveness and perturbed insulin secretion. | 23476019
 |
DNA methylation and histone modifications modulate the β1,3 galactosyltransferase β3Gal-T5 native promoter in cancer cells.
Anna Caretti,Silvia M Sirchia,Silvia Tabano,Aida Zulueta,Fabio Dall'olio,Marco Trinchera
The international journal of biochemistry & cell biology
44
2012
요약 표시
The native promoter of β1,3 galactosyltransferase β3Gal-T5 contributes to the expression of the enzyme and its oligosaccharide products, such as Lewis antigens, in many tissues. It is mainly sensitive to nuclear factor NF-Y and located nearby two CpG islands. To elucidate the regulation of the native promoter, we analyzed NF-Y protein and β3Gal-T5 mRNA, and found that NF-Y is scarcely modulated among various cell lines and biopsies from normal or cancerous colon. Conversely, β3Gal-T5 expression levels vary in the cell lines and are strongly down-regulated in colon cancer. We also performed quantitative methylation analysis of β3Gal-T5 CpG islands and found an inverse correlation between mRNA expression and DNA methylation. In particular, the methylation levels of both islands are always increased in cancer, with respect to the corresponding normal counterpart, in matched normal and tumor samples of colon and breast origin. Moreover, treatment with chromatin remodeling agents 5-aza-2'deoxycytidine and trichostatin A does not restore transcription in completely negative cells, but only increases expression in basally positive cells. However, methylation analysis after 5-aza-2'deoxycytidine treatment revealed partial demethylation of both islands in all treated cells. Finally, chromatin immunoprecipitation assays on β3Gal-T5 promoter showed that histone H3K4 trymethylation, H3K79 dimethylation, and H3K9-14 acetylation are high in cells expressing the transcript, and very low in those negative, while H4K20 trimethylation and H3K27 dimethylation are the opposite. We conclude that complex epigenetic modulation underlies the regulation of β3Gal-T5 native promoter. | 22001559
 |