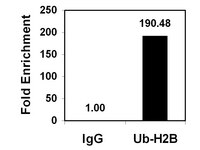
RE-IIBP Methylates H3K79 and Induces MEIS1-mediated Apoptosis via H2BK120 Ubiquitination by RNF20.
Woo Park, J; Kim, KB; Kim, JY; Chae, YC; Jeong, OS; Seo, SB
Scientific reports
5
12485
2015
概要を表示する
Histone lysine methylation contributes to transcriptional regulation by serving as a platform for the recruitment of various cofactors. Intense studies have been conducted for elucidating the functional meaning of H3K79 methylation, and to date, the only known HMTase responsible for the modification was DOT1L. In this study, we report that the MMSET isoform RE-IIBP has HMTase activity for H3K79. It was uncovered that RE-IIBP up-regulates MEIS1 transcription through H3K79 methylation via recruitment to the MEIS1 promoter. By means of proteomic and biochemical analysis, association of RE-IIBP with the E3 ubiquitin ligase RNF20 was demonstrated for synergistic activation of MEIS1 transcription via H3K79 HMTase activity. Furthermore, It was observed that RE-IIBP induces MEIS1-mediated apoptosis, which was dependent on H2BK120 ubiquitination by RNF20. These findings suggest RE-IIBP as another candidate for further studies to elucidate the mechanism of H3K79 methylation and its biological functions. | 26206755
 |
H2B monoubiquitylation is a 5'-enriched active transcription mark and correlates with exon-intron structure in human cells.
Jung, I; Kim, SK; Kim, M; Han, YM; Kim, YS; Kim, D; Lee, D
Genome research
22
1026-35
2012
概要を表示する
H2B monoubiquitylation (H2Bub1), which is required for multiple methylations of both H3K4 and H3K79, has been implicated in gene expression in numerous organisms ranging from yeast to human. However, the molecular crosstalk between H2Bub1 and other modifications, especially the methylations of H3K4 and H3K79, remains unclear in vertebrates. To better understand the functional role of H2Bub1, we measured genome-wide histone modification patterns in human cells. Our results suggest that H2Bub1 has dual roles, one that is H3 methylation dependent, and another that is H3 methylation independent. First, H2Bub1 is a 5'-enriched active transcription mark and co-occupies with H3K79 methylations in actively transcribed regions. Second, this study shows for the first time that H2Bub1 plays a histone H3 methylations-independent role in chromatin architecture. Furthermore, the results of this work indicate that H2Bub1 is largely positioned at the exon-intron boundaries of highly expressed exons, and it demonstrates increased occupancy in skipped exons compared with flanking exons in the human and mouse genomes. Our findings collectively suggest that a potentiating mechanism links H2Bub1 to both H3K79 methylations in actively transcribed regions and the exon-intron structure of highly expressed exons via the regulation of nucleosome dynamics during transcription elongation. | 22421545
 |
Reelin is a target of polyglutamine expanded ataxin-7 in human spinocerebellar ataxia type 7 (SCA7) astrocytes.
McCullough, SD; Xu, X; Dent, SY; Bekiranov, S; Roeder, RG; Grant, PA
Proceedings of the National Academy of Sciences of the United States of America
109
21319-24
2012
概要を表示する
Spinocerebellar ataxia type 7 (SCA7) is an autosomal-dominant neurodegenerative disorder that results from polyglutamine expansion of the ataxin-7 (ATXN7) protein. Remarkably, although mutant ATXN7 is expressed throughout the body, pathology is restricted primarily to the cerebellum and retina. One major goal has been to identify factors that contribute to the tissue specificity of SCA7. Here we describe the development and use of a human astrocyte cell culture model to identify reelin, a factor intimately involved in the development and maintenance of Purkinje cells and the cerebellum as a whole, as an ATXN7 target gene. We found that polyglutamine expansion decreased ATXN7 occupancy, which correlated with increased levels of histone H2B monoubiquitination, at the reelin promoter. Treatment with trichostatin A, but not other histone deacetylase inhibitors, partially restored reelin transcription and promoted the accumulation of mutant ATXN7 into nuclear inclusions. Our findings suggest that reelin could be a previously unknown factor involved in the tissue specificity of SCA7 and that trichostatin A may ameliorate deleterious effects of the mutant ATXN7 protein by promoting its sequestration away from promoters into nuclear inclusions. | 23236151
 |
Histone crosstalk directed by H2B ubiquitination is required for chromatin boundary integrity.
Ma, MK; Heath, C; Hair, A; West, AG
PLoS genetics
7
e1002175
2011
概要を表示する
Genomic maps of chromatin modifications have provided evidence for the partitioning of genomes into domains of distinct chromatin states, which assist coordinated gene regulation. The maintenance of chromatin domain integrity can require the setting of boundaries. The HS4 insulator element marks the 3' boundary of a heterochromatin region located upstream of the chicken β-globin gene cluster. Here we show that HS4 recruits the E3 ligase RNF20/BRE1A to mediate H2B mono-ubiquitination (H2Bub1) at this insulator. Knockdown experiments show that RNF20 is required for H2Bub1 and processive H3K4 methylation. Depletion of RNF20 results in a collapse of the active histone modification signature at the HS4 chromatin boundary, where H2Bub1, H3K4 methylation, and hyperacetylation of H3, H4, and H2A.Z are rapidly lost. A remarkably similar set of events occurs at the HSA/HSB regulatory elements of the FOLR1 gene, which mark the 5' boundary of the same heterochromatin region. We find that persistent H2Bub1 at the HSA/HSB and HS4 elements is required for chromatin boundary integrity. The loss of boundary function leads to the sequential spreading of H3K9me2, H3K9me3, and H4K20me3 over the entire 50 kb FOLR1 and β-globin region and silencing of FOLR1 expression. These findings show that the HSA/HSB and HS4 boundary elements direct a cascade of active histone modifications that defend the FOLR1 and β-globin gene loci from the pervasive encroachment of an adjacent heterochromatin domain. We propose that many gene loci employ H2Bub1-dependent boundaries to prevent heterochromatin spreading. | 21811414
 |
Elongin B-mediated epigenetic alteration of viral chromatin correlates with efficient human cytomegalovirus gene expression and replication.
Hwang, J; Saffert, RT; Kalejta, RF
mBio
2
e00023-11
2011
概要を表示する
Elongins B and C are members of complexes that increase the efficiency of transcriptional elongation by RNA polymerase II (RNAPII) and enhance the monoubiquitination of histone H2B, an epigenetic mark of actively transcribed genes. Here we show that, in addition to its role in facilitating transcription of the cellular genome, elongin B also enhances gene expression from the double-stranded DNA genome of human cytomegalovirus (HCMV), a pathogenic herpesvirus. Reducing the level of elongin B by small interfering RNA- or short hairpin RNA-mediated knockdown decreased viral mRNA expression, viral protein accumulation, viral DNA replication, and infectious virion production. Chromatin immunoprecipitation analysis indicated viral genome occupancy of the elongating form of RNAPII, and monoubiquitinated histone H2B was reduced in elongin B-deficient cells. These data suggest that, in addition to the previously documented epigenetic regulation of transcriptional initiation, HCMV also subverts cellular elongin B-mediated epigenetic mechanisms for enhancing transcriptional elongation to enhance viral gene expression and virus replication.The genetic and epigenetic control of transcription initiation at both cellular and viral promoters is well documented. Recently, the epigenetic modification of histone H2B monoubiquitination throughout the bodies of cellular genes has been shown to enhance the elongation of RNA polymerase II-initiated transcripts. Mechanisms that might control the elongation of viral transcripts are less well studied. Here we show that, as with cellular genes, elongin B-mediated monoubiquitination of histone H2B also facilitates the transcriptional elongation of human cytomegalovirus genes. This and perhaps other epigenetic markings of actively transcribed regions may help in identifying viral genes expressed during in vitro latency or during natural infections of humans. Furthermore, this work identifies a novel, tractable model system to further study the regulation of transcriptional elongation in living cells. 記事全文 | 21447700
 |