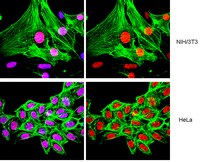
Modulation of chromatin remodelling induced by the freshwater cyanotoxin cylindrospermopsin in human intestinal caco-2 cells.
Huguet, A; Hatton, A; Villot, R; Quenault, H; Blanchard, Y; Fessard, V
PloS one
9
e99121
2014
Mostrar resumen
Cylindrospermopsin (CYN) is a cyanotoxin that has been recognised as an emerging potential public health risk. Although CYN toxicity has been demonstrated, the mechanisms involved have not been fully characterised. To identify some key pathways related to this toxicity, we studied the transcriptomic profile of human intestinal Caco-2 cells exposed to a sub-toxic concentration of CYN (1.6 µM for 24hrs) using a non-targeted approach. CYN was shown to modulate different biological functions which were related to growth arrest (with down-regulation of cdkn1a and uhrf1 genes), and DNA recombination and repair (with up-regulation of aptx and pms2 genes). Our main results reported an increased expression of some histone-modifying enzymes (histone acetyl and methyltransferases MYST1, KAT5 and EHMT2) involved in chromatin remodelling, which is essential for initiating transcription. We also detected greater levels of acetylated histone H2A (Lys5) and dimethylated histone H3 (Lys4), two products of these enzymes. In conclusion, CYN overexpressed proteins involved in DNA damage repair and transcription, including modifications of nucleosomal histones. Our results highlighted some new cell processes induced by CYN. | | 24921660
 |
Epigenetic chromatin modifications in barley after mutagenic treatment.
Braszewska-Zalewska, A; Tylikowska, M; Kwasniewska, J; Szymanowska-Pulka, J
Journal of applied genetics
55
449-56
2014
Mostrar resumen
In addition to their normal developmental processes, plants have evolved complex genetic and epigenetic regulatory mechanisms to cope with various environmental stresses. It has been shown that both DNA methylation and histone modifications are involved in DNA damage response to various types of stresses. In this study, we focused on the involvement of two mutagenic agents, chemical (maleic acid hydrazide; MH) and physical (gamma rays), on the global epigenetic modifications of chromatin in barley. Our results indicate that both mutagens strongly influence the level of histone methylation and acetylation. Moreover, we found that gamma irradiation, in contrast to MH, has a more robust influence on the DNA methylation level. This is the first study that brings together mutagenic treatment along with its impact at the level of epigenetic modifications examined using the immunohistochemical method. | | 24939040
 |
Spatial distribution of epigenetic modifications in Brachypodium distachyon embryos during seed maturation and germination.
Wolny, E; Braszewska-Zalewska, A; Hasterok, R
PloS one
9
e101246
2014
Mostrar resumen
Seed development involves a plethora of spatially and temporally synchronised genetic and epigenetic processes. Although it has been shown that epigenetic mechanisms, such as DNA methylation and chromatin remodelling, act on a large number of genes during seed development and germination, to date the global levels of histone modifications have not been studied in a tissue-specific manner in plant embryos. In this study we analysed the distribution of three epigenetic markers, i.e. H4K5ac, H3K4me2 and H3K4me1 in 'matured', 'dry' and 'germinating' embryos of a model grass, Brachypodium distachyon (Brachypodium). Our results indicate that the abundance of these modifications differs considerably in various organs and tissues of the three types of Brachypodium embryos. Embryos from matured seeds were characterised by the highest level of H4K5ac in RAM and epithelial cells of the scutellum, whereas this modification was not observed in the coleorhiza. In this type of embryos H3K4me2 was most evident in epithelial cells of the scutellum. In 'dry' embryos H4K5ac was highest in the coleorhiza but was not present in the nuclei of the scutellum. H3K4me1 was the most elevated in the coleoptile but absent from the coleorhiza, whereas H3K4me2 was the most prominent in leaf primordia and RAM. In embryos from germinating seeds H4K5ac was the most evident in the scutellum but not present in the coleoptile, similarly H3K4me1 was the highest in the scutellum and very low in the coleoptile, while the highest level of H3K4me2 was observed in the coleoptile and the lowest in the coleorhiza. The distinct patterns of epigenetic modifications that were observed may be involved in the switch of the gene expression profiles in specific organs of the developing embryo and may be linked with the physiological changes that accompany seed desiccation, imbibition and germination. | | 25006668
 |
Tissue-specific epigenetic modifications in root apical meristem cells of Hordeum vulgare.
Braszewska-Zalewska, AJ; Wolny, EA; Smialek, L; Hasterok, R
PloS one
8
e69204
2013
Mostrar resumen
Epigenetic modifications of chromatin structure are essential for many biological processes, including growth and reproduction. Patterns of DNA and histone modifications have recently been widely studied in many plant species, although there is virtually no data on the spatial and temporal distribution of epigenetic markers during plant development. Accordingly, we have used immunostaining techniques to investigate epigenetic modifications in the root apical meristem of Hordeum vulgare. Histone H4 acetylation (H4K5ac), histone H3 dimethylation (H3K4me2, H3K9me2) and DNA methylation (5mC) patterns were established for various root meristem tissues. Distinct levels of those modifications were visualised in the root cap, epidermis, cortex and vascular tissues. The lateral root cap cells seem to display the highest level of H3K9me2 and 5mC. In the epidermis, the highest level of 5mC and H3K9me2 was detected in the nuclei from the boundary of the proximal meristem and the elongation zone, while the vascular tissues were characterized by the highest level of H4K5ac. Some of the modified histones were also detectable in the cytoplasm in a highly tissue-specific manner. Immunolocalisation of epigenetic modifications of chromatin carried out in this way, on longitudinal or transverse sections, provides a unique topographic context within the organ, and will provide some answers to the significant biological question of tissue differentiation processes during root development in a monocotyledon plant species. | Immunofluorescence | 23935955
 |
LIV-1 ZIP ectodomain shedding in prion-infected mice resembles cellular response to transition metal starvation.
Sepehr Ehsani,Ashkan Salehzadeh,Hairu Huo,William Reginold,Cosmin L Pocanschi,Hezhen Ren,Hansen Wang,Kelvin So,Christine Sato,Mohadeseh Mehrabian,Robert Strome,William S Trimble,Lili-Naz Hazrati,Ekaterina Rogaeva,David Westaway,George A Carlson,Gerold Schmitt-Ulms
Journal of molecular biology
422
2011
Mostrar resumen
We recently documented the co-purification of members of the LIV-1 subfamily of ZIP (Zrt-, Irt-like Protein) zinc transporters (LZTs) with the cellular prion protein (PrP(C)) and, subsequently, established that the prion gene family descended from an ancestral LZT gene. Here, we begin to address whether the study of LZTs can shed light on the biology of prion proteins in health and disease. Starting from an observation of an abnormal LZT immunoreactive band in prion-infected mice, subsequent cell biological analyses uncovered a surprisingly coordinated biology of ZIP10 (an LZT member) and prion proteins that involves alterations to N-glycosylation and endoproteolysis in response to manipulations to the extracellular divalent cation milieu. Starving cells of manganese or zinc, but not copper, causes shedding of the N1 fragment of PrP(C) and of the ectodomain of ZIP10. For ZIP10, this posttranslational biology is influenced by an interaction between its PrP-like ectodomain and a conserved metal coordination site within its C-terminal multi-spanning transmembrane domain. The transition metal starvation-induced cleavage of ZIP10 can be differentiated by an immature N-glycosylation signature from a constitutive cleavage targeting the same site. Data from this work provide a first glimpse into a hitherto neglected molecular biology that ties PrP to its LZT cousins and suggest that manganese or zinc starvation may contribute to the etiology of prion disease in mice. | | 22687393
 |
RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation.
Carey, Michael, et al.
Mol. Cell, 24: 481-7 (2006)
2005
Mostrar resumen
The coordinated action of histone acetyltransferases (HATs) and ATP-dependent chromatin remodeling enzymes in promoter-dependent transcription initiation represents a paradigm for how epigenetic information regulates gene expression. However, little is known about how such enzymes function during transcription elongation. Here, we investigated the role of RSC, a bromodomain-containing ATPase, in nucleosome transcription in vitro. Purified S. cerevisiae RNA polymerase II (Pol II) arrests at two primary locations on a positioned mononucleosome. RSC stimulates passage of Pol II through these sites. The function of RSC in elongation requires the energy of ATP hydrolysis. Moreover, the SAGA and NuA4 HATs strongly stimulated RSC's effect on elongation. The stimulation correlates closely with acetyl-CoA-dependent recruitment of RSC to nucleosomes. Thus, RSC can recognize acetylated nucleosomes and facilitate passage of Pol II through them. These data support the view that histone modifications regulate accessibility of the coding region to Pol II. | | 17081996
 |
Acetylated histone H4 is reduced in human gastric adenomas and carcinomas
Ono, S, et al
J Exp Clin Cancer Res, 21:377-82 (2002)
2002
| | 12385581
 |