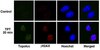
Splice variants of SmgGDS control small GTPase prenylation and membrane localization.
Berg, TJ; Gastonguay, AJ; Lorimer, EL; Kuhnmuench, JR; Li, R; Fields, AP; Williams, CL
The Journal of biological chemistry
285
35255-66
2010
Show Abstract
Ras and Rho small GTPases possessing a C-terminal polybasic region (PBR) are vital signaling proteins whose misregulation can lead to cancer. Signaling by these proteins depends on their ability to bind guanine nucleotides and their prenylation with a geranylgeranyl or farnesyl isoprenoid moiety and subsequent trafficking to cellular membranes. There is little previous evidence that cellular signals can restrain nonprenylated GTPases from entering the prenylation pathway, leading to the general belief that PBR-possessing GTPases are prenylated as soon as they are synthesized. Here, we present evidence that challenges this belief. We demonstrate that insertion of the dominant negative mutation to inhibit GDP/GTP exchange diminishes prenylation of Rap1A and RhoA, enhances prenylation of Rac1, and does not detectably alter prenylation of K-Ras. Our results indicate that the entrance and passage of these small GTPases through the prenylation pathway is regulated by two splice variants of SmgGDS, a protein that has been reported to promote GDP/GTP exchange by PBR-possessing GTPases and to be up-regulated in several forms of cancer. We show that the previously characterized 558-residue SmgGDS splice variant (SmgGDS-558) selectively associates with prenylated small GTPases and facilitates trafficking of Rap1A to the plasma membrane, whereas the less well characterized 607-residue SmgGDS splice variant (SmgGDS-607) associates with nonprenylated GTPases and regulates the entry of Rap1A, RhoA, and Rac1 into the prenylation pathway. These results indicate that guanine nucleotide exchange and interactions with SmgGDS splice variants can regulate the entrance and passage of PBR-possessing small GTPases through the prenylation pathway. Full Text Article | 20709748
 |
Mechanisms underlying expression of Tn10 encoded tetracycline resistance.
Hillen, W and Berens, C
Annu. Rev. Microbiol., 48: 345-69 (1994)
1994
Show Abstract
Tetracycline-resistance determinants encoding active efflux of the drug are widely distributed in gram-negative bacteria and unique with respect to genetic organization and regulation of expression. Each determinant consists of two genes called tetA and tetR, which are oriented with divergent polarity, and between them is a central regulatory region with overlapping promoters and operators. The amino acid sequences of the encoded proteins are 43-78% identical. The resistance protein TetA is a tetracycline/metal-proton antiporter located in the cytoplasmic membrane, while the regulatory protein TetR is a tetracycline inducible repressor. TetR binds via a helix-turn-helix motif to the two tet operators, resulting in repression of both genes. A detailed model of the repressor-operator complex has been proposed on the basis of biochemical and genetic data. The tet genes are differentially regulated so that repressor synthesis can occur before the resistance protein is expressed. This has been demonstrated for the Tn10-encoded tet genes and may be a common property of all tet determinants, as suggested by the similar locations of operators with respect to promoters. Induction is mediated by a tetracycline-metal complex and requires only nanomolar concentrations of the drug. This is the most sensitive effector-inducible system of transcriptional regulation known to date. The crystal structure of the TetR-tetracycline/metal complex shows the Tet repressor in the induced, non-DNA binding conformation. The structural interpretation of many noninducible TetR mutants has offered insight into the conformational changes associated with the switch between inducing and repressing structures of TetR. Tc is buried in the core of TetR, where it is held in place by multiple contacts to the protein. | 7826010
 |