A co-localization model of paired ChIP-seq data using a large ENCODE data set enables comparison of multiple samples.
Maehara, Kazumitsu, et al.
Nucleic Acids Res., 41: 54-62 (2013)
2013
Kivonat megmutatása
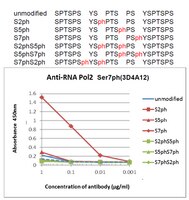
Deep sequencing approaches, such as chromatin immunoprecipitation by sequencing (ChIP-seq), have been successful in detecting transcription factor-binding sites and histone modification in the whole genome. An approach for comparing two different ChIP-seq data would be beneficial for predicting unknown functions of a factor. We propose a model to represent co-localization of two different ChIP-seq data. We showed that a meaningful overlapping signal and a meaningless background signal can be separated by this model. We applied this model to compare ChIP-seq data of RNA polymerase II C-terminal domain (CTD) serine 2 phosphorylation with a large amount of peak-called data, including ChIP-seq and other deep sequencing data in the Encyclopedia of DNA Elements (ENCODE) project, and then extracted factors that were related to RNA polymerase II CTD serine 2 in HeLa cells. We further analyzed RNA polymerase II CTD serine 7 phosphorylation, of which their function is still unclear in HeLa cells. Our results were characterized by the similarity of localization for transcription factor/histone modification in the ENCODE data set, and this suggests that our model is appropriate for understanding ChIP-seq data for factors where their function is unknown. | 23125363
 |
β-Catenin signaling is a critical event in ErbB2-mediated mammary tumor progression.
Schade, Babette, et al.
Cancer Res., 73: 4474-87 (2013)
2013
Kivonat megmutatása
Although ERBB2 amplification and overexpression is correlated with poor outcome in breast cancer, the molecular mechanisms underlying the aggressive nature of these tumors has not been fully elucidated. To investigate this further, we have used a transgenic mouse model of ErbB2-driven tumor progression (ErbB2(KI) model) that recapitulates clinically relevant events, including selective amplification of the core erbB2 amplicon. By comparing the transcriptional profiles of ErbB2(KI) mammary tumors and human ERBB2-positive breast cancers, we show that ErbB2(KI) tumors possess molecular features of the basal subtype of ERBB2-positive human breast cancer, including activation of canonical β-catenin signaling. Inhibition of β-catenin-dependent signaling in ErbB2(KI)-derived tumor cells using RNA interference impaired tumor initiation and metastasis. Furthermore, treatment of ErbB2(KI) or human ERBB2-overexpressing tumor cells with a selective β-catenin/CBP inhibitor significantly decreased proliferation and ErbB2 expression. Collectively, our data indicate that ERBB2-mediated breast cancer progression requires β-catenin signaling and can be therapeutically targeted by selective β-catenin/CBP inhibitors. | 23720052
 |