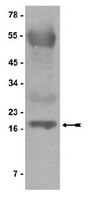
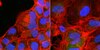
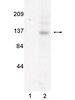
Herpes Simplex Virus 1 ICP4 Forms Complexes with TFIID and Mediator in Virus-Infected Cells.
Lester, Jonathan T and Deluca, Neal A
J. Virol., 85: 5733-44 (2011)
2011
Show Abstract
The infected cell polypeptide 4 (ICP4) of herpes simplex virus 1 (HSV-1) is a regulator of viral transcription that is required for productive infection. Since viral genes are transcribed by cellular RNA polymerase II (RNA pol II), ICP4 must interact with components of the pol II machinery to regulate viral gene expression. It has been shown previously that ICP4 interacts with TATA box-binding protein (TBP), TFIIB, and the TBP-associated factor 1 (TAF1) in vitro. In this study, ICP4-containing complexes were isolated from infected cells by tandem affinity purification (TAP). Forty-six proteins that copurified with ICP4 were identified by mass spectrometry. Additional copurifying proteins were identified by Western blot analysis. These included 11 components of TFIID and 4 components of the Mediator complex. The significance of the ICP4-Mediator interaction was further investigated using immunofluorescence and chromatin immunoprecipitation. Mediator was found to colocalize with ICP4 starting at early and continuing into late times of infection. In addition, Mediator was recruited to viral promoters in an ICP4-dependent manner. Taken together, the data suggest that ICP4 interacts with components of TFIID and Mediator in the context of viral infection, and this may explain the broad transactivation properties of ICP4. | | 21450820
 |
Functional analysis of epigenetic regulation of tandem RhopH1/clag genes reveals a role in Plasmodium falciparum growth.
Comeaux, CA; Coleman, BI; Bei, AK; Whitehurst, N; Duraisingh, MT
Molecular microbiology
80
378-90
2011
Show Abstract
The Plasmodium RhopH complex is a high molecular weight antigenic complex consisting of three subunits - RhopH1/clag, RhopH2 and RhopH3 - located in the rhoptry secretory organelles of the invasive merozoite. In Plasmodium falciparum RhopH1/clag is encoded by one of five clag genes. Two highly similar paralogous genes, clag 3.1 and clag 3.2, are mutually exclusively expressed. Here we show clonal switching from the clag 3.2 to the clag 3.1 paralogue in vitro. Chromatin immunoprecitation studies suggest that silencing of either clag 3 paralogue is associated with the enrichment of specific histone modifications associated with heterochromatin. We were able to disrupt the clag 3.2 gene, with a drug cassette inserted into the clag 3.2 locus being readily silenced in a position-dependent and sequence-independent manner. Activation of this drug cassette by drug selection results in parasites with the clag 3.1 locus silenced and lack full-length clag 3.1 or 3.2 transcripts. These clag 3-null parasites demonstrate a significant growth inhibition compared with wild-type parasites, providing the first genetic evidence for a role for these proteins in efficient parasite proliferation. Epigenetic regulation of these chromosomally proximal members of a multigene family provides a mechanism for both immune evasion and functional diversification. | Immunoprecipitation | 21320181
 |
Interaction between Hhex and SOX13 Modulates Wnt/TCF Activity.
Marfil V, Moya M, Pierreux CE, Castell JV, Lemaigre FP, Real FX, Bort R
The Journal of biological chemistry
285
5726-37
2010
Show Abstract
Fine-tuning of the Wnt/TCF pathway is crucial for multiple embryological processes, including liver development. Here we describe how the interaction between Hhex (hematopoietically expressed homeobox) and SOX13 (SRY-related high mobility group box transcription factor 13), modulates Wnt/TCF pathway activity. Hhex is a homeodomain factor expressed in multiple endoderm-derived tissues, like the liver, where it is essential for proper development. The pleiotropic expression of Hhex during embryonic development and its dual role as a transcriptional repressor and activator suggest the presence of different tissue-specific partners capable of modulating its activity and function. While searching for developmentally regulated Hhex partners, we set up a yeast two-hybrid screening using an E9.5-10.5 mouse embryo library and the N-terminal domain of Hhex as bait. Among the putative protein interactors, we selected SOX13 for further characterization. We found that SOX13 interacts directly with full-length Hhex, and we delineated the interaction domains within the two proteins. SOX13 is known to repress Wnt/TCF signaling by interacting with TCF1. We show that Hhex is able to block the SOX13-dependent repression of Wnt/TCF activity by displacing SOX13 from the SOX13.TCF1 complex. Moreover, Hhex de-repressed the Wnt/TCF pathway in the ventral foregut endoderm of cultured mouse embryos electroporated with a SOX13-expressing plasmid. We conclude that the interaction between Hhex and SOX13 may contribute to control Wnt/TCF signaling in the early embryo. Full Text Article | | 20028982
 |
MUC1 regulates nuclear localization and function of the epidermal growth factor receptor.
Bitler, BG; Goverdhan, A; Schroeder, JA
J Cell Sci
123
1716-23
2010
Show Abstract
Alteration of protein trafficking and localization is associated with several diseases, including cystic fibrosis, breast cancer, colorectal cancer, leukemia and diabetes. Specifically, aberrant nuclear localization of the epidermal growth factor receptor (EGFR), a receptor tyrosine kinase, is a poor prognostic indicator in several epithelial carcinomas. It is now appreciated that in addition to signaling from the plasma membrane, EGFR also trafficks to the nucleus, and can directly bind the promoter regions of genes encoding cyclin D1 (CCND1) and B-Myb (MYBL2). We have previously established that loss of MUC1 in an EGFR-dependent transgenic mouse model of breast cancer correlates with the loss of cyclin D1 expression. Here, we provide evidence for a novel regulatory function of MUC1 in the trafficking and nuclear activity of EGFR. We found that MUC1 and EGFR interact in the nucleus of breast cancer cells, which promotes the accumulation of chromatin-bound EGFR. Additionally, the presence of MUC1 results in significant colocalization of EGFR and phosphorylated RNA polymerase II, indicating that MUC1 influences the association of EGFR with transcriptionally active promoter regions. Importantly, we found that the loss of MUC1 expression resulted in a decrease in the interaction between EGFR and the CCND1 promoter, which translated to a significant decrease in cyclin D1 protein expression. This data offers insights into a novel regulatory mechanism of EGFR nuclear function and could have important implications for evaluating nuclear localization in cancer. Full Text Article | | 20406885
 |
Ikaros is a regulator of Il10 expression in CD4+ T cells.
Umetsu, SE; Winandy, S
J Immunol
183
5518-25
2009
Show Abstract
IL-10 is a regulatory cytokine critical for controlling inflammatory responses. Here we show that Ikaros, a zinc finger DNA-binding protein, plays an important role in the regulation of Il10 in murine CD4(+) T cells. Upon initial stimulation of the TCR, T cells deficient in Ikaros express significantly lower levels of IL-10 compared with wild-type T cells. In addition, under Th2 skewing conditions, which induce IL-10 production by wild-type T cells, Ikaros null T cells are unable to properly differentiate, producing only low levels of IL-10. Expression of a dominant-negative isoform of Ikaros in wild-type Th2 cells represses IL-10 production but does not significantly alter expression levels of the genes encoding the transcription factors GATA-3 and T-bet. Furthermore, expression of Ikaros in Ikaros null T cells restores expression of the Th2 cytokines IL-10 and IL-4 while reducing production of the Th1 cytokine, IFN-gamma. Coexpression of Ikaros and GATA-3 further increases IL-10 production, showing that these two factors have an additive effect on activating Il10 expression. Finally, we show that Ikaros binds to conserved regulatory regions of the Il10 gene locus in Th2 cells, supporting a direct role for Ikaros in Il10 expression. Thus, we provide evidence for Ikaros as a regulator of Il10 and Ifng gene expression and suggest a role for Ikaros in directing lineage-specific cytokine gene activation and repression. Full Text Article | | 19828627
 |
Transcriptional regulation of CD4 gene expression by T cell factor-1/beta-catenin pathway.
Zhaofeng Huang, Huimin Xie, Vassilio Ioannidis, Werner Held, Hans Clevers, Maureen S Sadim, Zuoming Sun
Journal of immunology (Baltimore, Md. : 1950)
176
4880-7
2006
Show Abstract
By interacting with MHC class II molecules, CD4 facilitates lineage development as well as activation of Th cells. Expression of physiological levels of CD4 requires a proximal CD4 enhancer to stimulate basic CD4 promoter activity. T cell factor (TCF)-1/beta-catenin pathway has previously been shown to regulate thymocyte survival via up-regulating antiapoptotic molecule Bcl-xL. By both loss and gain of function studies, in this study we show additional function of TCF-1/beta-catenin pathway in the regulation of CD4 expression in vivo. Mice deficient in TCF-1 displayed significantly reduced protein and mRNA levels of CD4 in CD4+ CD8+ double-positive (DP) thymocytes. A transgene encoding Bcl-2 restored survival but not CD4 levels of TCF-1(-/-) DP cells. Thus, TCF-1-regulated survival and CD4 expression are two separate events. In contrast, CD4 levels were restored on DP TCF-1(-/-) cells by transgenic expression of a wild-type TCF-1, but not a truncated TCF-1 that lacks a domain required for interacting with beta-catenin. Furthermore, forced expression of a stabilized beta-catenin, a coactivator of TCF-1, resulted in up-regulation of CD4. TCF-1 or stabilized beta-catenin greatly stimulated activity of a CD4 reporter gene driven by a basic CD4 promoter and the CD4 enhancer. However, mutation of a potential TCF binding site located within the enhancer abrogated TCF-1 and beta-catenin-mediated activation of CD4 reporter. Finally, recruitment of TCF-1 to CD4 enhancer was detected in wild-type but not TCF-1 null mice by chromatin-immunoprecipitation analysis. Thus, our results demonstrated that TCF/beta-catenin pathway enhances CD4 expression in vivo by recruiting TCF-1 to stimulate CD4 enhancer activity. | | 16585583
 |