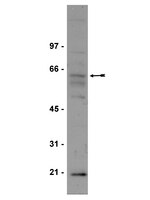
Inhibition of insulin-like growth factor receptor/AKT/mammalian target of rapamycin axis targets colorectal cancer stem cells by attenuating mevalonate-isoprenoid pathway in vitro and in vivo.
Sharon, C; Baranwal, S; Patel, NJ; Rodriguez-Agudo, D; Pandak, WM; Majumdar, AP; Krystal, G; Patel, BB
Oncotarget
6
15332-47
2015
Abstract anzeigen
We observed a co-upregulation of the insulin-like growth factor receptor (IGF-1R)/AKT/mammalian target of rapamycin (mTOR) [InAT] axis and the mevalonate-isoprenoid biosynthesis (MIB) pathways in colorectal cancer stem cells (CSCs) in an unbiased approach. Hence, we hypothesized that the InAT axis might regulate the MIB pathway to govern colorectal CSCs growth. Stimulation (IGF-1) or inhibition (IGF-1R depletion and pharmacological inhibition of IGF-1R/mTOR) of the InAT axis produced induction or attenuation of CSC growth as well as expression of CSC markers and self-renewal factors respectively. Intriguingly, activation of the InAT axis (IGF-1) caused significant upregulation of the MIB pathway genes (both mRNA and protein); while its inhibition produced the opposite effects in colonospheres. More importantly, supplementation with dimethylallyl- and farnesyl-PP, MIB metabolites downstream of isopentenyl-diphosphate delta isomerase (IDI), but not mevalonate and isopentenyl-pp that are upstream of IDI, resulted in a near-complete reversal of the suppressive effect of the InAT axis inhibitors on CSCs growth. The latter findings suggest a specific regulation of the MIB pathway by the InAT axis distal to the target of statins that inhibit 3-hydroxy-3-methyl-glutaryl-CoA reductase (HMGCR). Effects of IGF-1R inhibition on colonic CSCs proliferation and the MIB pathway were confirmed in an 'in vivo' HCT-116 xenograft model. These observations establish a novel mechanistic link between the InAT axis that is commonly deregulated in colorectal cancer and the MIB pathway in regulation of colonic CSCs growth. Hence, the InAT-MIB corridor is a novel target for developing paradigm shifting optimum anti-CSCs therapies for colorectal cancer. | | | 25895029
 |
CUL4A contributes to the biology of basal-like breast tumors through modulation of cell growth and antitumor immune response.
Saucedo-Cuevas, LP; Ruppen, I; Ximénez-Embún, P; Domingo, S; Gayarre, J; Muñoz, J; Silva, JM; García, MJ; Benítez, J
Oncotarget
5
2330-43
2014
Abstract anzeigen
The CUL4A E3 ubiquitin ligase is involved in the regulation of many cellular processes and its amplification and/or overexpression has been observed in breast cancer. The 13q34 amplification, which is associated with the basal-like breast cancer subtype, has been proposed as one of the mechanism behind CUL4A up-regulation. However, the specific contribution of CUL4A to the biology of basal-like breast tumors has not yet been elucidated. In this work, by using cellular models of basal phenotype, we show the inhibitory effect of CUL4A silencing in the proliferation and growth of breast cancer cells both, in vitro and in vivo. We also demonstrate the transforming capacity of CUL4A exogenous overexpression in the 184B5 human mammary epithelial cells in vitro. Our results suggest a synergistic effect between CUL4A high levels and the activation of the RAS pathway in the tumorigenesis of basal-like breast cancer tumors. In addition, by using a proteomics approach we have defined novel candidate proteins and pathways that might mediate the oncogenic effect of CUL4A. In particular, we report a putative role of CUL4A in bypassing the immune system in breast cancer through the down-regulation of several molecules involved in the immune surveillance. These findings provide insight into the oncogenic properties of CUL4A in basal-like breast cancer and highlight the therapeutic opportunities to target CUL4A. | | | 24870930
 |
Oncogenic protein MTBP interacts with MYC to promote tumorigenesis.
Grieb, BC; Gramling, MW; Arrate, MP; Chen, X; Beauparlant, SL; Haines, DS; Xiao, H; Eischen, CM
Cancer research
74
3591-602
2014
Abstract anzeigen
Despite its involvement in most human cancers, MYC continues to pose a challenge as a readily tractable therapeutic target. Here we identify the MYC transcriptional cofactors TIP48 and TIP49 and MYC as novel binding partners of Mdm2-binding protein (MTBP), a functionally undefined protein that we show is oncogenic and overexpressed in many human cancers. MTBP associated with MYC at promoters and increased MYC-mediated transcription, proliferation, neoplastic transformation, and tumor development. In breast cancer specimens, we determined overexpression of both MYC and MTBP was associated with a reduction in 10-year patient survival compared with MYC overexpression alone. MTBP was also frequently co-amplified with MYC in many human cancers. Mechanistic investigations implicated associations with TIP48/TIP49 as well as MYC in MTBP function in cellular transformation and the growth of human breast cancer cells. Taken together, our findings show MTBP functions with MYC to promote malignancy, identifying this protein as a novel general therapeutic target in human cancer. | | | 24786788
 |
Rapid identification of regulatory microRNAs by miTRAP (miRNA trapping by RNA in vitro affinity purification).
Braun, J; Misiak, D; Busch, B; Krohn, K; Hüttelmaier, S
Nucleic acids research
42
e66
2014
Abstract anzeigen
MicroRNAs (miRNAs) control gene expression at the post-transcriptional level. However, the identification of miRNAs regulating the fate of a specific messenger RNA remains limited due to the imperfect complementarity of miRNAs and targeted transcripts. Here, we describe miTRAP (miRNA trapping by RNA in vitro affinity purification), an advanced protocol of previously reported MS2-tethering approaches. MiTRAP allows the rapid identification of miRNAs targeting an in vitro transcribed RNA in cell lysates. Selective co-purification of regulatory miRNAs was confirmed for the MYC- as well as ZEB2-3'UTR, two well-established miRNA targets in vivo. Combined with miRNA-sequencing, miTRAP identified in addition to miRNAs reported to control MYC expression, 18 novel candidates including not in silico predictable miRNAs. The evaluation of 10 novel candidate miRNAs confirmed 3'UTR-dependent regulation of MYC expression as well as putative non-canonical targeting sites for the not in silico predictable candidates. In conclusion, miTRAP provides a rapid, cost-effective and easy-to-handle protocol allowing the identification of regulatory miRNAs for RNAs of choice in a cellular context of interest. Most notably, miTRAP not only identifies in silico predictable but also unpredictable miRNAs regulating the expression of a specific target RNA. | Western Blotting | | 24510096
 |
Identification of c-MYC SUMOylation by mass spectrometry.
Kalkat, M; Chan, PK; Wasylishen, AR; Srikumar, T; Kim, SS; Ponzielli, R; Bazett-Jones, DP; Raught, B; Penn, LZ
PloS one
9
e115337
2014
Abstract anzeigen
The c-MYC transcription factor is a master regulator of many cellular processes and deregulation of this oncogene has been linked to more than 50% of all cancers. This deregulation can take many forms, including altered post-translational regulation. Here, using immunoprecipitation combined with mass spectrometry, we identified a MYC SUMOylation site (K326). Abrogation of signaling through this residue by substitution with arginine (K326R) has no obvious effects on MYC half-life, intracellular localization, transcriptional targets, nor on the biological effects of MYC overexpression in two different cell systems assessed for soft agar colony formation, proliferation, and apoptosis. While we have definitively demonstrated that MYC SUMOylation can occur on K326, future work will be needed to elucidate the mechanisms and biological significance of MYC regulation by SUMOylation. | Western Blotting | | 25522242
 |
A mouse model for inducible overexpression of Prdm14 results in rapid-onset and highly penetrant T-cell acute lymphoblastic leukemia (T-ALL).
Carofino, BL; Ayanga, B; Justice, MJ
Disease models & mechanisms
6
1494-506
2013
Abstract anzeigen
PRDM14 functions in embryonic stem cell (ESC) maintenance to promote the expression of pluripotency-associated genes while suppressing differentiation genes. Expression of PRDM14 is tightly regulated and typically limited to ESCs and primordial germ cells; however, aberrant expression is associated with tumor initiation in a wide variety of human cancers, including breast cancer and leukemia. Here, we describe the generation of a Cre-recombinase-inducible mouse model for the spatial and temporal control of Prdm14 misexpression [ROSA26 floxed-stop Prdm14 (R26PR)]. When R26PR is mated to either of two Cre lines, Mx1-cre or MMTV-cre, mice develop early-onset T-cell acute lymphoblastic leukemia (T-ALL) with median overall survival of 41 and 64 days for R26PR;Mx1-cre and R26PR;MMTV-cre, respectively. T-ALL is characterized by the accumulation of immature single-positive CD8 cells and their widespread infiltration. Leukemia is preceded by a dramatic expansion of cells resembling hematopoietic stem cells and lymphoid-committed progenitors prior to disease onset, accompanied by a blockage in B-cell differentiation at the early pro-B stage. Rapid-onset PRDM14-induced T-ALL requires factors that are present in stem and progenitor cells: R26PR;dLck-cre animals, which express Prdm14 starting at the double-positive stage of thymocyte development, do not develop disease. PRDM14-induced leukemic cells contain high levels of activated NOTCH1 and downstream NOTCH1 targets, including MYC and HES1, and are sensitive to pharmacological inhibition of NOTCH1 with the γ-secretase inhibitor DAPT. Greater than 50% of human T-ALLs harbor activating mutations in NOTCH1; thus, our model carries clinically relevant molecular aberrations. The penetrance, short latency and involvement of the NOTCH1 pathway will make this hematopoietic R26PR mouse model ideal for future studies on disease initiation, relapse and novel therapeutic drug combinations. Furthermore, breeding R26PR to additional Cre lines will allow for the continued development of novel cancer models. | | | 24046360
 |
Suppression of Ras/Mapk pathway signaling inhibits Myc-induced lymphomagenesis.
Gramling, MW; Eischen, CM
Cell death and differentiation
19
1220-7
2011
Abstract anzeigen
Although the Myc transcription factor has been shown necessary for the oncogenic function of Ras, the contribution of Ras pathway signaling to the oncogenic function of Myc remains unresolved. We report the novel findings that Myc alone induced Ras/Mapk pathway signaling, and increased signaling following growth factor stimulation. Deletion of the scaffold protein kinase suppressor of Ras 1 (Ksr1) attenuated signaling through the Ras/Mapk pathway, including activation following Myc induction. B cells that lacked Ksr1 exhibited reduced proliferation and increased cytokine deprivation-induced apoptosis. Overexpression of Myc rescued the proliferation defect of Ksr1-null B cells, but loss of Ksr1 increased sensitivity of B cells to Myc-induced apoptosis. Notably, there was a significant delay in lymphoma development in Ksr1-null mice overexpressing Myc in B cells (Eμ-myc transgenic mice). There was an elevated frequency of p53 inactivation, indicative of increased selective pressure to bypass the p53 tumor suppressor pathway, in Ksr1-null Eμ-myc lymphomas. Therefore, loss of Ksr1 inhibits Ras/Mapk pathway signaling leading to increased Myc-induced B-cell apoptosis, and this results in reduced B-cell transformation and lymphoma development. Our data indicate that suppression of Myc-induced Ras/Mapk pathway signaling significantly impairs Myc oncogenic function. These results fill a significant gap in knowledge about Myc and should open new avenues of therapeutic intervention for Myc-overexpressing malignancies. | | | 22301919
 |
Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling.
Wong, VW; Stange, DE; Page, ME; Buczacki, S; Wabik, A; Itami, S; van de Wetering, M; Poulsom, R; Wright, NA; Trotter, MW; Watt, FM; Winton, DJ; Clevers, H; Jensen, KB
Nature cell biology
14
401-8
2011
Abstract anzeigen
Maintenance of adult tissues is carried out by stem cells and is sustained throughout life in a highly ordered manner. Homeostasis within the stem-cell compartment is governed by positive- and negative-feedback regulation of instructive extrinsic and intrinsic signals. ErbB signalling is a prerequisite for maintenance of the intestinal epithelium following injury and tumour formation. As ErbB-family ligands and receptors are highly expressed within the stem-cell niche, we hypothesize that strong endogenous regulators must control the pathway in the stem-cell compartment. Here we show that Lrig1, a negative-feedback regulator of the ErbB receptor family, is highly expressed by intestinal stem cells and controls the size of the intestinal stem-cell niche by regulating the amplitude of growth-factor signalling. Intestinal stem-cell maintenance has so far been attributed to a combination of Wnt and Notch activation and Bmpr inhibition. Our findings reveal ErbB activation as a strong inductive signal for stem-cell proliferation. This has implications for our understanding of ErbB signalling in tissue development and maintenance and the progression of malignant disease. | | Mouse | 22388892
 |
Determination of protein interactome of transcription factor Sox2 in embryonic stem cells engineered for inducible expression of four reprogramming factors.
Gao, Z; Cox, JL; Gilmore, JM; Ormsbee, BD; Mallanna, SK; Washburn, MP; Rizzino, A
The Journal of biological chemistry
287
11384-97
2011
Abstract anzeigen
Unbiased proteomic screens provide a powerful tool for defining protein-protein interaction networks. Previous studies employed multidimensional protein identification technology to identify the Sox2-interactome in embryonic stem cells (ESC) undergoing differentiation in response to a small increase in the expression of epitope-tagged Sox2. Thus far the Sox2-interactome in ESC has not been determined. To identify the Sox2-interactome in ESC, we engineered ESC for inducible expression of different combinations of epitope-tagged Sox2 along with Oct4, Klf4, and c-Myc. Epitope-tagged Sox2 was used to circumvent the lack of suitable Sox2 antibodies needed to perform an unbiased proteomic screen of Sox2-associated proteins. Although i-OS-ESC differentiate when both Oct4 and Sox2 are elevated, i-OSKM-ESC do not differentiate even when the levels of the four transcription factors are coordinately elevated ∼2-3-fold. Our findings with i-OS-ESC and i-OSKM-ESC provide new insights into the reasons why ESC undergo differentiation when Sox2 and Oct4 are elevated in ESC. Importantly, the use of i-OSKM-ESC enabled us to identify the Sox2-interactome in undifferentiated ESC. Using multidimensional protein identification technology, we identified greater than 70 proteins that associate with Sox2 in ESC. We extended these findings by testing the function of the Sox2-assoicated protein Smarcd1 and demonstrate that knockdown of Smarcd1 disrupts the self-renewal of ESC and induces their differentiation. Together, our work provides the first description of the Sox2-interactome in ESC and indicates that Sox2 along with other master regulators is part of a highly integrated protein-protein interaction landscape in ESC. | | | 22334693
 |
Downregulation of HuR as a new mechanism of doxorubicin resistance in breast cancer cells.
Latorre, E; Tebaldi, T; Viero, G; Spartà, AM; Quattrone, A; Provenzani, A
Molecular cancer
11
13
2011
Abstract anzeigen
HuR, an RNA binding protein involved in the post-transcriptional regulation of a wide spectrum of mRNAs, has been demonstrated to be a determinant of carcinogenesis and tumor aggressiveness in several cancer types. In this study, we investigated the role of HuR in the apoptosis and in the chemoresistance induced by the widely used anticancer drug doxorubicin in human breast cancer cells (MCF-7).We showed that HuR acts in the early phase of cell response to doxorubicin, being induced to translocate into the cytoplasm upon phosphorylation. Reducing HuR levels diminished the apoptotic response to doxorubicin. Doxorubicin-induced apoptosis was also correlated with the presence of HuR in the cytoplasm. Rottlerin, which was able to block HuR nuclear export, had correspondingly antagonistic effects with doxorubicin on cell toxicity. The proapoptotic activity of HuR was not due to cleavage to an active form, as was previously reported. In in vitro selected doxorubicin resistant MCF-7 cells (MCF-7/doxoR) overexpressing the multidrug resistance (MDR) related ABCG2 transporter, we observed a significant HuR downregulation that was paralleled by a corresponding downregulation of HuR targets and by loss of rottlerin toxicity. Restoration of HuR expression in these cells resensitized MCF-7/doxoR cells to doxorubicin, reactivating the apoptotic response.The present study shows that HuR is necessary to elicit the apoptotic cell response to doxorubicin and that restoration of HuR expression in resistant cells resensitizes them to the action of this drug, thereby identifying HuR as a key protein in doxorubicin pharmacology. | | | 22436134
 |