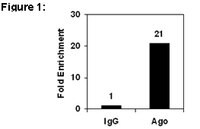
A high throughput experimental approach to identify miRNA targets in human cells.
Tan, Lu Ping, et al.
Nucleic Acids Res., 37: e137 (2009)
2009
Zobrazit abstrakt
The study of human microRNAs is seriously hampered by the lack of proper tools allowing genome-wide identification of miRNA targets. We performed Ribonucleoprotein ImmunoPrecipitation-gene Chip (RIP-Chip) using antibodies against wild-type human Ago2 in untreated Hodgkin lymphoma (HL) cell lines. Ten to thirty percent of the gene transcripts from the genome were enriched in the Ago2-IP fraction of untreated cells, representing the HL miRNA-targetome. In silico analysis indicated that approximately 40% of these gene transcripts represent targets of the abundantly co-expressed miRNAs. To identify targets of miR-17/20/93/106, RIP-Chip with anti-miR-17/20/93/106 treated cells was performed and 1189 gene transcripts were identified. These genes were analyzed for miR-17/20/93/106 target sites in the 5'-UTRs, coding regions and 3'-UTRs. Fifty-one percent of them had miR-17/20/93/106 target sites in the 3'-UTR while 19% of them were predicted miR-17/20/93/106 targets by TargetScan. Luciferase reporter assay confirmed targeting of miR-17/20/93/106 to the 3'-UTRs of 8 out of 10 genes. In conclusion, we report a method which can establish the miRNA-targetome in untreated human cells and identify miRNA specific targets in a high throughput manner. This approach is applicable to identify miRNA targets in any human tissue sample or purified cell population in an unbiased and physiologically relevant manner. | 19734348
 |
Involvement of AGO1 and AGO2 in mammalian transcriptional silencing.
Janowski, BA; Huffman, KE; Schwartz, JC; Ram, R; Nordsell, R; Shames, DS; Minna, JD; Corey, DR
Nat Struct Mol Biol
13
787-92
2005
Zobrazit abstrakt
Duplex RNAs complementary to messenger RNA inhibit translation in mammalian cells by RNA interference (RNAi). Studies have reported that RNAs complementary to promoter DNA also inhibit gene expression. Here we show that the human homologs of Argonaute-1 (AGO1) and Argonaute-2 (AGO2) link the silencing pathways that target mRNA with pathways mediating recognition of DNA. We find that synthetic antigene RNAs (agRNAs) complementary to transcription start sites or more upstream regions of gene promoters inhibit gene transcription. This silencing occurs in the nucleus, requires high promoter activity and does not necessarily require histone modification. AGO1 and AGO2 associate with promoter DNA in cells treated with agRNAs, and inhibiting expression of AGO1 or AGO2 reverses transcriptional and post-transcriptional silencing. Our data indicate key linkages and important mechanistic distinctions between transcriptional and post-transcriptional silencing pathways in mammalian cells. | 16936728
 |
Comparative expression analysis of the MAGED genes during embryogenesis and brain development.
Mathieu Bertrand,Ivo Huijbers,Patrick Chomez,Olivier De Backer
Developmental dynamics : an official publication of the American Association of Anatomists
230
2004
Zobrazit abstrakt
The MAGED gene subfamily contains three genes in mouse and four in human. The MAGED1, D2, and D3 proteins are highly conserved between mouse and human, whereas paralogues are less conserved between each other. This finding suggests that each MAGED protein exerts a distinct function. To get a better insight into their physiological roles, we have analyzed their expression patterns during embryogenesis and brain development. In the mouse, Maged3 expression is restricted to the central nervous system where it was mostly detected in postmitotic neurons. Maged2 is mainly expressed in tissues of mesodermal origin. The expression pattern of Maged1 roughly summarizes that of Maged2 and Maged3; however, contrary to that of Maged3, it includes the proliferative zones of the nervous system. We observed a discrepancy between Maged1 expression levels of RNA and protein, suggesting that its expression is regulated at a posttranscriptional level during the mouse development. | 15162511
 |