Alpha7 nicotinic acetylcholine receptor is required for blood-brain barrier injury-related CNS disorders caused by Cryptococcus neoformans and HIV-1 associated comorbidity factors.
Zhang, B; Yu, JY; Liu, LQ; Peng, L; Chi, F; Wu, CH; Jong, A; Wang, SF; Cao, H; Huang, SH
BMC infectious diseases
15
352
2015
Mostra il sommario
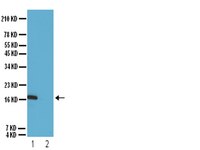
Cryptococcal meningitis is the most common fungal infection of the central nervous system (CNS) in HIV/AIDS. HIV-1 virotoxins (e.g., gp41) are able to induce disorders of the blood-brain barrier (BBB), which mainly consists of BMEC. Our recent study suggests that α7 nAChR is an essential regulator of inflammation, which contributes to regulation of NF-κB signaling, neuroinflammation and BBB disorders caused by microbial (e.g., HIV-1 gp120) and non-microbial [e.g., methamphetamine (METH)] factors. However, the underlying mechanisms for multiple comorbidities are unclear.In this report, an aggravating role of α7 nAChR in host defense against CNS disorders caused by these comorbidities was demonstrated by chemical [inhibitor: methyllycaconitine (MLA)] and genetic (α7(-/-) mice) blockages of α7 nAChR.As shown in our in vivo studies, BBB injury was significantly reduced in α7(-/-) mice infected with C. neoformans. Stimulation by the gp41 ectodomain peptide (gp41-I90) and METH was abolished in the α7(-/-) animals. C. neoformans and gp41-I90 could activate NF-κB. Gp41-I90- and METH-induced monocyte transmigration and senescence were significantly inhibited by MLA and CAPE (caffeic acid phenethyl ester, an NF-κB inhibitor).Collectively, our data suggest that α7 nAChR plays a detrimental role in the host defense against C. neoformans- and HIV-1 associated comorbidity factors-induced BBB injury and CNS disorders. | | | 26285576
 |
Methylation of histone H3 lysine 9 occurs during translation.
Rivera, C; Saavedra, F; Alvarez, F; Díaz-Celis, C; Ugalde, V; Li, J; Forné, I; Gurard-Levin, ZA; Almouzni, G; Imhof, A; Loyola, A
Nucleic acids research
43
9097-106
2015
Mostra il sommario
Histone post-translational modifications are key contributors to chromatin structure and function, and participate in the maintenance of genome stability. Understanding the establishment and maintenance of these marks, along with their misregulation in pathologies is thus a major focus in the field. While we have learned a great deal about the enzymes regulating histone modifications on nucleosomal histones, much less is known about the mechanisms establishing modifications on soluble newly synthesized histones. This includes methylation of lysine 9 on histone H3 (H3K9), a mark that primes the formation of heterochromatin, a critical chromatin landmark for genome stability. Here, we report that H3K9 mono- and dimethylation is imposed during translation by the methyltransferase SetDB1. We discuss the importance of these results in the context of heterochromatin establishment and maintenance and new therapeutic opportunities in pathologies where heterochromatin is perturbed. | | | 26405197
 |
The potential of GMP-compliant platelet lysate to induce a permissive state for cardiovascular transdifferentiation in human mediastinal adipose tissue-derived mesenchymal stem cells.
Siciliano, C; Chimenti, I; Bordin, A; Ponti, D; Iudicone, P; Peruzzi, M; Rendina, EA; Calogero, A; Pierelli, L; Ibrahim, M; De Falco, E
BioMed research international
2015
162439
2015
Mostra il sommario
Human adipose tissue-derived mesenchymal stem cells (ADMSCs) are considered eligible candidates for cardiovascular stem cell therapy applications due to their cardiac transdifferentiation potential and immunotolerance. Over the years, the in vitro culture of ADMSCs by platelet lysate (PL), a hemoderivate containing numerous growth factors and cytokines derived from platelet pools, has allowed achieving a safe and reproducible methodology to obtain high cell yield prior to clinical administration. Nevertheless, the biological properties of PL are still to be fully elucidated. In this brief report we show the potential ability of PL to induce a permissive state of cardiac-like transdifferentiation and to cause epigenetic modifications. RTPCR results indicate an upregulation of Cx43, SMA, c-kit, and Thy-1 confirmed by immunofluorescence staining, compared to standard cultures with foetal bovine serum. Moreover, PL-cultured ADMSCs exhibit a remarkable increase of both acetylated histones 3 and 4, with a patient-dependent time trend, and methylation at lysine 9 on histone 3 preceding the acetylation. Expression levels of p300 and SIRT-1, two major regulators of histone 3, are also upregulated after treatment with PL. In conclusion, PL could unravel novel biological properties beyond its routine employment in noncardiac applications, providing new insights into the plasticity of human ADMSCs. | | | 26495284
 |
Analysis of Histones H3 and H4 Reveals Novel and Conserved Post-Translational Modifications in Sugarcane.
Moraes, I; Yuan, ZF; Liu, S; Souza, GM; Garcia, BA; Casas-Mollano, JA
PloS one
10
e0134586
2015
Mostra il sommario
Histones are the main structural components of the nucleosome, hence targets of many regulatory proteins that mediate processes involving changes in chromatin. The functional outcome of many pathways is "written" in the histones in the form of post-translational modifications that determine the final gene expression readout. As a result, modifications, alone or in combination, are important determinants of chromatin states. Histone modifications are accomplished by the addition of different chemical groups such as methyl, acetyl and phosphate. Thus, identifying and characterizing these modifications and the proteins related to them is the initial step to understanding the mechanisms of gene regulation and in the future may even provide tools for breeding programs. Several studies over the past years have contributed to increase our knowledge of epigenetic gene regulation in model organisms like Arabidopsis, yet this field remains relatively unexplored in crops. In this study we identified and initially characterized histones H3 and H4 in the monocot crop sugarcane. We discovered a number of histone genes by searching the sugarcane ESTs database. The proteins encoded correspond to canonical histones, and their variants. We also purified bulk histones and used them to map post-translational modifications in the histones H3 and H4 using mass spectrometry. Several modifications conserved in other plants, and also novel modified residues, were identified. In particular, we report O-acetylation of serine, threonine and tyrosine, a recently identified modification conserved in several eukaryotes. Additionally, the sub-nuclear localization of some well-studied modifications (i.e., H3K4me3, H3K9me2, H3K27me3, H3K9ac, H3T3ph) is described and compared to other plant species. To our knowledge, this is the first report of histones H3 and H4 as well as their post-translational modifications in sugarcane, and will provide a starting point for the study of chromatin regulation in this crop. | | | 26226299
 |
Hierarchical clustering of breast cancer methylomes revealed differentially methylated and expressed breast cancer genes.
Lin, IH; Chen, DT; Chang, YF; Lee, YL; Su, CH; Cheng, C; Tsai, YC; Ng, SC; Chen, HT; Lee, MC; Chen, HW; Suen, SH; Chen, YC; Liu, TT; Chang, CH; Hsu, MT
PloS one
10
e0118453
2015
Mostra il sommario
Oncogenic transformation of normal cells often involves epigenetic alterations, including histone modification and DNA methylation. We conducted whole-genome bisulfite sequencing to determine the DNA methylomes of normal breast, fibroadenoma, invasive ductal carcinomas and MCF7. The emergence, disappearance, expansion and contraction of kilobase-sized hypomethylated regions (HMRs) and the hypomethylation of the megabase-sized partially methylated domains (PMDs) are the major forms of methylation changes observed in breast tumor samples. Hierarchical clustering of HMR revealed tumor-specific hypermethylated clusters and differential methylated enhancers specific to normal or breast cancer cell lines. Joint analysis of gene expression and DNA methylation data of normal breast and breast cancer cells identified differentially methylated and expressed genes associated with breast and/or ovarian cancers in cancer-specific HMR clusters. Furthermore, aberrant patterns of X-chromosome inactivation (XCI) was found in breast cancer cell lines as well as breast tumor samples in the TCGA BRCA (breast invasive carcinoma) dataset. They were characterized with differentially hypermethylated XIST promoter, reduced expression of XIST, and over-expression of hypomethylated X-linked genes. High expressions of these genes were significantly associated with lower survival rates in breast cancer patients. Comprehensive analysis of the normal and breast tumor methylomes suggests selective targeting of DNA methylation changes during breast cancer progression. The weak causal relationship between DNA methylation and gene expression observed in this study is evident of more complex role of DNA methylation in the regulation of gene expression in human epigenetics that deserves further investigation. | | | 25706888
 |
The preRC protein ORCA organizes heterochromatin by assembling histone H3 lysine 9 methyltransferases on chromatin.
Giri, S; Aggarwal, V; Pontis, J; Shen, Z; Chakraborty, A; Khan, A; Mizzen, C; Prasanth, KV; Ait-Si-Ali, S; Ha, T; Prasanth, SG
eLife
4
2015
Mostra il sommario
Heterochromatic domains are enriched with repressive histone marks, including histone H3 lysine 9 methylation, written by lysine methyltransferases (KMTs). The pre-replication complex protein, origin recognition complex-associated (ORCA/LRWD1), preferentially localizes to heterochromatic regions in post-replicated cells. Its role in heterochromatin organization remained elusive. ORCA recognizes methylated H3K9 marks and interacts with repressive KMTs, including G9a/GLP and Suv39H1 in a chromatin context-dependent manner. Single-molecule pull-down assays demonstrate that ORCA-ORC (Origin Recognition Complex) and multiple H3K9 KMTs exist in a single complex and that ORCA stabilizes H3K9 KMT complex. Cells lacking ORCA show alterations in chromatin architecture, with significantly reduced H3K9 di- and tri-methylation at specific chromatin sites. Changes in heterochromatin structure due to loss of ORCA affect replication timing, preferentially at the late-replicating regions. We demonstrate that ORCA acts as a scaffold for the establishment of H3K9 KMT complex and its association and activity at specific chromatin sites is crucial for the organization of heterochromatin structure. | | | 25922909
 |
An H2A histone isotype regulates estrogen receptor target genes by mediating enhancer-promoter-3'-UTR interactions in breast cancer cells.
Su, CH; Tzeng, TY; Cheng, C; Hsu, MT
Nucleic acids research
42
3073-88
2014
Mostra il sommario
A replication-dependent histone H2A isotype, H2ac, is upregulated in MCF-7 cells and in estrogen receptor-positive clinical breast cancer tissues. Cellular depletion of this H2A isotype leads to defective estrogen signaling, loss of cell proliferation and cell cycle arrest at G0/G1 phase. H2ac mediates regulation of estrogen receptor target genes, particularly BCL2 and c-MYC, by recruiting estrogen receptor alpha through its HAR domain and facilitating the formation of a chromatin loop between the promoter, enhancer and 3'-untranslated region of the respective genes. These findings reveal a new role for histone isotypes in the regulation of gene expression in cancer cells, and suggest that these molecules may be targeted for anti-cancer drug discovery. | | | 24371278
 |
The interaction of MYC with the trithorax protein ASH2L promotes gene transcription by regulating H3K27 modification.
Ullius, A; Lüscher-Firzlaff, J; Costa, IG; Walsemann, G; Forst, AH; Gusmao, EG; Kapelle, K; Kleine, H; Kremmer, E; Vervoorts, J; Lüscher, B
Nucleic acids research
42
6901-20
2014
Mostra il sommario
The appropriate expression of the roughly 30,000 human genes requires multiple layers of control. The oncoprotein MYC, a transcriptional regulator, contributes to many of the identified control mechanisms, including the regulation of chromatin, RNA polymerases, and RNA processing. Moreover, MYC recruits core histone-modifying enzymes to DNA. We identified an additional transcriptional cofactor complex that interacts with MYC and that is important for gene transcription. We found that the trithorax protein ASH2L and MYC interact directly in vitro and co-localize in cells and on chromatin. ASH2L is a core subunit of KMT2 methyltransferase complexes that target histone H3 lysine 4 (H3K4), a mark associated with open chromatin. Indeed, MYC associates with H3K4 methyltransferase activity, dependent on the presence of ASH2L. MYC does not regulate this methyltransferase activity but stimulates demethylation and subsequently acetylation of H3K27. KMT2 complexes have been reported to associate with histone H3K27-specific demethylases, while CBP/p300, which interact with MYC, acetylate H3K27. Finally WDR5, another core subunit of KMT2 complexes, also binds directly to MYC and in genome-wide analyses MYC and WDR5 are associated with transcribed promoters. Thus, our findings suggest that MYC and ASH2L-KMT2 complexes cooperate in gene transcription by controlling H3K27 modifications and thereby regulate bivalent chromatin. | | | 24782528
 |
Epigenetic chromatin modifications in barley after mutagenic treatment.
Braszewska-Zalewska, A; Tylikowska, M; Kwasniewska, J; Szymanowska-Pulka, J
Journal of applied genetics
55
449-56
2014
Mostra il sommario
In addition to their normal developmental processes, plants have evolved complex genetic and epigenetic regulatory mechanisms to cope with various environmental stresses. It has been shown that both DNA methylation and histone modifications are involved in DNA damage response to various types of stresses. In this study, we focused on the involvement of two mutagenic agents, chemical (maleic acid hydrazide; MH) and physical (gamma rays), on the global epigenetic modifications of chromatin in barley. Our results indicate that both mutagens strongly influence the level of histone methylation and acetylation. Moreover, we found that gamma irradiation, in contrast to MH, has a more robust influence on the DNA methylation level. This is the first study that brings together mutagenic treatment along with its impact at the level of epigenetic modifications examined using the immunohistochemical method. | | | 24939040
 |
Cerebellar oxidative DNA damage and altered DNA methylation in the BTBR T+tf/J mouse model of autism and similarities with human post mortem cerebellum.
Shpyleva, S; Ivanovsky, S; de Conti, A; Melnyk, S; Tryndyak, V; Beland, FA; James, SJ; Pogribny, IP
PloS one
9
e113712
2014
Mostra il sommario
The molecular pathogenesis of autism is complex and involves numerous genomic, epigenomic, proteomic, metabolic, and physiological alterations. Elucidating and understanding the molecular processes underlying the pathogenesis of autism is critical for effective clinical management and prevention of this disorder. The goal of this study is to investigate key molecular alterations postulated to play a role in autism and their role in the pathophysiology of autism. In this study we demonstrate that DNA isolated from the cerebellum of BTBR T+tf/J mice, a relevant mouse model of autism, and from human post-mortem cerebellum of individuals with autism, are both characterized by an increased levels of 8-oxo-7-hydrodeoxyguanosine (8-oxodG), 5-methylcytosine (5mC), and 5-hydroxymethylcytosine (5hmC). The increase in 8-oxodG and 5mC content was associated with a markedly reduced expression of the 8-oxoguanine DNA-glycosylase 1 (Ogg1) and increased expression of de novo DNA methyltransferases 3a and 3b (Dnmt3a and Dnmt3b). Interestingly, a rise in the level of 5hmC occurred without changes in the expression of ten-eleven translocation expression 1 (Tet1) and Tet2 genes, but significantly correlated with the presence of 8-oxodG in DNA. This finding and similar elevation in 8-oxodG in cerebellum of individuals with autism and in the BTBR T+tf/J mouse model warrant future large-scale studies to specifically address the role of OGG1 alterations in pathogenesis of autism. | Western Blotting | | 25423485
 |