A signature motif mediating selective interactions of BCL11A with the NR2E/F subfamily of orphan nuclear receptors.
Chan, CM; Fulton, J; Montiel-Duarte, C; Collins, HM; Bharti, N; Wadelin, FR; Moran, PM; Mongan, NP; Heery, DM
Nucleic acids research
41
9663-79
2013
Abstract anzeigen
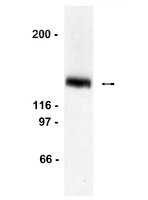
Despite their physiological importance, selective interactions between nuclear receptors (NRs) and their cofactors are poorly understood. Here, we describe a novel signature motif (F/YSXXLXXL/Y) in the developmental regulator BCL11A that facilitates its selective interaction with members of the NR2E/F subfamily. Two copies of this motif (named here as RID1 and RID2) permit BCL11A to bind COUP-TFs (NR2F1;NR2F2;NR2F6) and Tailless/TLX (NR2E1), whereas RID1, but not RID2, binds PNR (NR2E3). We confirmed the existence of endogenous BCL11A/TLX complexes in mouse cortex tissue. No interactions of RID1 and RID2 with 20 other ligand-binding domains from different NR subtypes were observed. We show that RID1 and RID2 are required for BCL11A-mediated repression of endogenous γ-globin gene and the regulatory non-coding transcript Bgl3, and we identify COUP-TFII binding sites within the Bgl3 locus. In addition to their importance for BCL11A function, we show that F/YSXXLXXL/Y motifs are conserved in other NR cofactors. A single FSXXLXXL motif in the NR-binding SET domain protein NSD1 facilitates its interactions with the NR2E/F subfamily. However, the NSD1 motif incorporates features of both LXXLL and FSXXLXXL motifs, giving it a distinct NR-binding pattern in contrast to other cofactors. In summary, our results provide new insights into the selectivity of NR/cofactor complex formation. | Western Blotting | 23975195
 |
PRAME is a golgi-targeted protein that associates with the Elongin BC complex and is upregulated by interferon-gamma and bacterial PAMPs.
Wadelin, FR; Fulton, J; Collins, HM; Tertipis, N; Bottley, A; Spriggs, KA; Falcone, FH; Heery, DM
PloS one
8
e58052
2013
Abstract anzeigen
Preferentially expressed antigen in melanoma (PRAME) has been described as a cancer-testis antigen and is associated with leukaemias and solid tumours. Here we show that PRAME gene transcription in leukaemic cell lines is rapidly induced by exposure of cells to bacterial PAMPs (pathogen associated molecular patterns) in combination with type 2 interferon (IFNγ). Treatment of HL60 cells with lipopolysaccharide or peptidoglycan in combination with IFNγ resulted in a rapid and transient induction of PRAME transcription, and increased association of PRAME transcripts with polysomes. Moreover, treatment with PAMPs/IFNγ also modulated the subcellular localisation of PRAME proteins in HL60 and U937 cells, resulting in targeting of cytoplasmic PRAME to the Golgi. Affinity purification studies revealed that PRAME associates with Elongin B and Elongin C, components of Cullin E3 ubiquitin ligase complexes. This occurs via direct interaction of PRAME with Elongin C, and PRAME colocalises with Elongins in the Golgi after PAMP/IFNγ treatment. PRAME was also found to co-immunoprecipitate core histones, consistent with its partial localisation to the nucleus, and was found to bind directly to histone H3 in vitro. Thus, PRAME is upregulated by signalling pathways that are activated in response to infection/inflammation, and its product may have dual functions as a histone-binding protein, and in directing ubiquitylation of target proteins for processing in the Golgi. | Western Blotting | 23460923
 |
Notch signaling is antagonized by SAO-1, a novel GYF-domain protein that interacts with the E3 ubiquitin ligase SEL-10 in Caenorhabditis elegans.
Hale, VA; Guiney, EL; Goldberg, LY; Haduong, JH; Kwartler, CS; Scangos, KW; Goutte, C
Genetics
190
1043-57
2011
Abstract anzeigen
Notch signaling pathways can be regulated through a variety of cellular mechanisms, and genetically compromised systems provide useful platforms from which to search for the responsible modulators. The Caenorhabditis elegans gene aph-1 encodes a component of γ-secretase, which is essential for Notch signaling events throughout development. By looking for suppressors of the incompletely penetrant aph-1(zu147) mutation, we identify a new gene, sao-1 (suppressor of aph-one), that negatively regulates aph-1(zu147) activity in the early embryo. The sao-1 gene encodes a novel protein that contains a GYF protein-protein interaction domain and interacts specifically with SEL-10, an Fbw7 component of SCF E3 ubiquitin ligases. We demonstrate that the embryonic lethality of aph-1(zu147) mutants can be suppressed by removing sao-1 activity or by mutations that disrupt the SAO-1-SEL-10 protein interaction. Decreased sao-1 activity also influences Notch signaling events when they are compromised at different molecular steps of the pathway, such as at the level of the Notch receptor GLP-1 or the downstream transcription factor LAG-1. Combined analysis of the SAO-1-SEL-10 protein interaction and comparisons of sao-1 and sel-10 genetic interactions suggest a possible role for SAO-1 as an accessory protein that participates with SEL-10 in downregulation of Notch signaling. This work provides the first mutant analysis of a GYF-domain protein in either C. elegans or Drosophila and introduces a new type of Fbw7-interacting protein that acts in a subset of Fbw7 functions. | | 22209900
 |
DNA replication origin function is promoted by H3K4 di-methylation in Saccharomyces cerevisiae.
Rizzardi, LF; Dorn, ES; Strahl, BD; Cook, JG
Genetics
192
371-84
2011
Abstract anzeigen
DNA replication is a highly regulated process that is initiated from replication origins, but the elements of chromatin structure that contribute to origin activity have not been fully elucidated. To identify histone post-translational modifications important for DNA replication, we initiated a genetic screen to identify interactions between genes encoding chromatin-modifying enzymes and those encoding proteins required for origin function in the budding yeast Saccharomyces cerevisiae. We found that enzymes required for histone H3K4 methylation, both the histone methyltransferase Set1 and the E3 ubiquitin ligase Bre1, are required for robust growth of several hypomorphic replication mutants, including cdc6-1. Consistent with a role for these enzymes in DNA replication, we found that both Set1 and Bre1 are required for efficient minichromosome maintenance. These phenotypes are recapitulated in yeast strains bearing mutations in the histone substrates (H3K4 and H2BK123). Set1 functions as part of the COMPASS complex to mono-, di-, and tri-methylate H3K4. By analyzing strains lacking specific COMPASS complex members or containing H2B mutations that differentially affect H3K4 methylation states, we determined that these replication defects were due to loss of H3K4 di-methylation. Furthermore, histone H3K4 di-methylation is enriched at chromosomal origins. These data suggest that H3K4 di-methylation is necessary and sufficient for normal origin function. We propose that histone H3K4 di-methylation functions in concert with other histone post-translational modifications to support robust genome duplication. | | 22851644
 |
Suppression and recovery of BRCA1-mediated transcription by HP1γ via modulation of promoter occupancy.
Choi, JD; Park, MA; Lee, JS
Nucleic acids research
40
11321-38
2011
Abstract anzeigen
Heterochromatin protein 1γ (HP1γ) is a chromatin protein involved in gene silencing. Herein, we show that HP1γ interacts with breast cancer type 1 susceptibility protein (BRCA1) and regulates BRCA1-mediated transcription via modulation of promoter occupancy and histone modification. We used several HP1γ mutants and small interfering RNAs for histone methyltransferases to show that BRCA1-HP1γ interaction, but not methylated histone binding, is important in HP1γ repression of BRCA1-mediated transcription. Time-lapse studies on promoter association and histone methylation after DNA damage revealed that HP1γ accumulates at the promoter before DNA damage, but BRCA1 is recruited at the promoter after the damage while promoter-resident HP1γ is disassembled. Importantly, HP1γ assembly recovers after release from the damage in a BRCA1-HP1γ interaction-dependent manner and targets SUV39H1. HP1γ/SUV39H1 restoration at the promoter results in BRCA1 disassembly and histone methylation, after which transcription repression resumes. We propose that through interaction with BRCA1, HP1γ is guided to the BRCA1 target promoter during recovery and functions in the activation-repression switch and recovery from BRCA1-mediated transcription in response to DNA damage. | | 23074186
 |
Direct identification of insulator components by insertional chromatin immunoprecipitation.
Fujita, T; Fujii, H
PloS one
6
e26109
2010
Abstract anzeigen
Comprehensive understanding of mechanisms of epigenetic regulation requires identification of molecules bound to genomic regions of interest in vivo. However, non-biased methods to identify molecules bound to specific genomic loci in vivo are limited. Here, we applied insertional chromatin immunoprecipitation (iChIP) to direct identification of components of insulator complexes, which function as boundaries of chromatin domain. We found that the chicken β-globin HS4 (cHS4) insulator complex contains an RNA helicase protein, p68/DDX5; an RNA species, steroid receptor RNA activator 1; and a nuclear matrix protein, Matrin-3, in vivo. Binding of p68 and Matrin-3 to the cHS4 insulator core sequence was mediated by CCCTC-binding factor (CTCF). Thus, our results showed that it is feasible to directly identify proteins and RNA bound to a specific genomic region in vivo by using iChIP. Volltextartikel | | 22043306
 |
Insertional chromatin immunoprecipitation: a method for isolating specific genomic regions.
Hoshino, A; Fujii, H
J Biosci Bioeng
108
446-9
2009
Abstract anzeigen
We established a novel method, insertional chromatin immunoprecipitation (iChIP), for isolation of specific genomic regions. In iChIP, specific genomic domains are immunoprecipitated with antibody against a tag, which is fused to the DNA-binding domain of an exogenous DNA-binding protein, whose recognition sequence is inserted into the genomic domains of interest. The iChIP method will be a useful tool for dissecting chromatin structure of genomic region of interest. | | 19804873
 |
Lack of nuclear translocation of cytoplasmic domains of IL-2/IL-15 receptor subunits.
Hodaka Fujii,Akemi Hoshino
Cytokine
46
2009
Abstract anzeigen
Some sensors of extracellular signaling molecules such as Notch and sterol response element binding protein (SREBP) receive ligand-induced intra-membrane proteolysis followed by nuclear translocation of their cytoplasmic domains to regulate gene expression programs in the nucleus. It has not been extensively examined whether ligand-induced intra-membrane proteolysis of type I cytokine receptors and nuclear translocation of cytoplasmic domains occur. Here, by using a sensitive reporter system, we examined this possibility for the interleukin-2 (IL-2) receptor (IL-2R) beta-chain (IL-2R beta) and the IL-15 receptor (IL-15R) alpha-chain (IL-15R alpha). Flowcytometric analysis revealed that ligand stimulation does not induce nuclear translocation of their cytoplasmic domains. In addition, overexpression of the cytoplasmic domain of the common cytokine receptor gamma-chain (gamma c) in an IL-2R-reconstituted Ba/F3-derived cell line did not affect any biological responses including cell survival, disproving potential roles of the cleaved cytoplasmic domain of gamma c as a signal transducer. Collectively, these results indicated that potential nuclear function of cleaved type I cytokine receptor subunits is not plausible. Volltextartikel | | 19329337
 |
Genome-wide analysis of the functions of a conserved surface on the corepressor Tup1.
Green, SR; Johnson, AD
Molecular biology of the cell
16
2605-13
2004
Abstract anzeigen
The general transcriptional repressor Tup1 is responsible for the regulation of a large, diverse set of genes in Saccharomyces cerevisiae, and functional homologues of Tup1 have been identified in many metazoans. The crystal structure for the C-terminal portion of Tup1 has been solved and, when sequences of Tup1 homologues from fungi and metazoans were compared, a highly conserved surface was revealed. In this article, we analyze five point mutations that lie on this conserved surface. A statistical analysis of expression microarrays demonstrates that the mutant alleles are deficient in the repression of different subsets of Tup1-regulated genes. We were able to rank the mutant alleles of TUP1 based on the severity of their repression defects measured both by the number of genes derepressed and by the magnitude of that derepression. For one particular class of genes, the mutations on the conserved surface disrupted recruitment of Tup1 to the repressed promoters. However, for the majority of the genes derepressed by the Tup1 point mutants, recruitment of Tup1 to the regulated promoters is largely unaffected. These mutations affect the mechanism of repression subsequent to recruitment of the complex and likely represent a disruption of a mechanism that is conserved in fungi and metazoans. This work demonstrates that the evolutionarily conserved surface of Tup1 interacts with two separate types of proteins-sequence-specific DNA-binding proteins responsible for recruiting Tup1 to promoters as well as components that are likely to function in a conserved repression mechanism. | | 15788561
 |
Tyrosine residues direct the ubiquitination and degradation of the NY-1 hantavirus G1 cytoplasmic tail.
Erika Geimonen, Imelyn Fernandez, Irina N Gavrilovskaya, Erich R Mackow
Journal of virology
77
10760-868
2003
Abstract anzeigen
The hantavirus G1 protein contains a long C-terminal cytoplasmic tail of 142 residues. Hantavirus pulmonary syndrome-associated hantaviruses contain conserved tyrosine residues near the C terminus of G1 which form an immunoreceptor tyrosine activation motif (ITAM) and interact with Src and Syk family kinases. During studies of the G1 ITAM we observed that fusion proteins containing the G1 cytoplasmic tail were poorly expressed. Expression of G1 cytoplasmic tail constructs were dramatically enhanced by treating cells with the proteasome inhibitor ALLN, suggesting that the protein is ubiquitinated and degraded via the 26S proteasome. By using a 6-His-tagged ubiquitin, we demonstrated that the G1 cytoplasmic tail is polyubiquitinated and degraded in the absence of proteasome inhibitors. Expression of only the ITAM-containing domain also directed protein ubiquitination and degradation in the absence of upstream residues. Deleting the C-terminal 51 residues of G1, including the ITAM, stabilized G1 and blocked polyubiquitination and degradation of the protein. Site-directed mutagenesis of both ITAM tyrosines (Y619 and Y632) to phenylalanine also blocked polyubiquitination of G1 proteins and dramatically enhanced G1 protein stability. In contrast, the presence of Y627, which is not part of the ITAM motif, had no effect on G1 stability. Mutagenesis of just Y619 enhanced G1 stability, inhibited G1 ubiquitination, and increased the half-life of G1 by threefold. Mutating only Y632 had less of an effect on G1 protein stability, although Y619 and Y632 synergistically contributed to G1 instability. These findings suggest that Y619, which is conserved in all hantaviruses, is the primary signal for directing G1 ubiquitination and degradation. Collectively these findings indicate that specific conserved tyrosines within the G1 cytoplasmic tail direct the polyubiquitination and degradation of expressed G1 proteins and provide a potential means for down-regulating hantavirus G1 surface glycoproteins and cellular proteins that interact with G1. Volltextartikel | | 14512526
 |